Omega-3 Fatty Acids Correlate with Gut Microbiome Diversity And
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Effect of Peptide Histidine Isoleucine on Water and Electrolyte Transport in the Human Jejunum
Gut: first published as 10.1136/gut.25.6.624 on 1 June 1984. Downloaded from Gut, 1984, 25, 624-628 Alimentary tract and pancreas Effect of peptide histidine isoleucine on water and electrolyte transport in the human jejunum K J MORIARTY, J E HEGARTY, K TATEMOTO, V MUTT, N D CHRISTOFIDES, S R BLOOM, AND J R WOOD From the Department of Gastroenterology, St Bartholomew's Hospital, London, The Liver Unit, King's College Hospital, London, Department ofMedicine, Hammersmith Hospital, London, and Department of Biochemistry, Karolinska Institute, Stockholm, Sweden SUMMARY Peptide histidine isoleucine, a 27 amino acid peptide with close amino acid sequence homology to vasoactive intestinal peptide and secretin, is distributed throughout the mammalian intestinal tract, where it has been localised to intramural neurones. An intestinal perfusion technique has been used to study the effect of intravenous peptide histidine isoleucine (44.5 pmol/kg/min) on water and electrolyte transport from a plasma like electrolyte solution in human jejunum in vivo. Peptide histidine isoleucine infusion produced peak plasma peptide histidine isoleucine concentrations in the range 2000-3000 pmolIl, flushing, tachycardia and a reduction in diastolic blood pressure. Peptide histidine isoleucine caused a significant inhibition of net absorption of water, sodium, potassium and bicarbonate and induced a net secretion of chloride, these changes being completely reversed during the post-peptide histidine isoleucine period. These findings suggest that endogenous peptide histidine isoleucine may participate in the neurohumoral regulation of water and electrolyte transport in the human jejunum. http://gut.bmj.com/ Peptide histidine isoleucine, isolated originally from INTESTINAL PERFUSION mammalian small intestine, is a 27-amino acid After an eight hour fast, each subject swallowed a peptide having close amino acid sequence homology double lumen intestinal perfusion tube, incorpo- to vasoactive intestinal peptide and secretin. -

Measurement of Metabolite Variations and Analysis of Related Gene Expression in Chinese Liquorice (Glycyrrhiza Uralensis) Plants
www.nature.com/scientificreports OPEN Measurement of metabolite variations and analysis of related gene expression in Chinese liquorice Received: 15 March 2017 Accepted: 28 March 2018 (Glycyrrhiza uralensis) plants under Published: xx xx xxxx UV-B irradiation Xiao Zhang1,2, Xiaoli Ding3,4, Yaxi Ji1,2, Shouchuang Wang5, Yingying Chen1,2, Jie Luo5, Yingbai Shen1,2 & Li Peng3,4 Plants respond to UV-B irradiation (280–315 nm wavelength) via elaborate metabolic regulatory mechanisms that help them adapt to this stress. To investigate the metabolic response of the medicinal herb Chinese liquorice (Glycyrrhiza uralensis) to UV-B irradiation, we performed liquid chromatography tandem mass spectrometry (LC-MS/MS)-based metabolomic analysis, combined with analysis of diferentially expressed genes in the leaves of plants exposed to UV-B irradiation at various time points. Fifty-four metabolites, primarily amino acids and favonoids, exhibited changes in levels after the UV-B treatment. The amino acid metabolism was altered by UV-B irradiation: the Asp family pathway was activated and closely correlated to Glu. Some amino acids appeared to be converted into antioxidants such as γ-aminobutyric acid and glutathione. Hierarchical clustering analysis revealed that various favonoids with characteristic groups were induced by UV-B. In particular, the levels of some ortho- dihydroxylated B-ring favonoids, which might function as scavengers of reactive oxygen species, increased in response to UV-B treatment. In general, unigenes encoding key enzymes involved in amino acid metabolism and favonoid biosynthesis were upregulated by UV-B irradiation. These fndings lay the foundation for further analysis of the mechanism underlying the response of G. -

Isoleucine, an Essential Amino Acid, Prevents Liver Metastases of Colon Cancer by Antiangiogenesis Kazumoto Murata1 and Masami Moriyama2
Research Article Isoleucine, an Essential Amino Acid, Prevents Liver Metastases of Colon Cancer by Antiangiogenesis Kazumoto Murata1 and Masami Moriyama2 1The First Department of Internal Medicine, Mie University School of Medicine, Tsu, Mie, Japan and 2Microbiology and Immunology, Keio University School of Medicine, Tokyo, Japan Abstract infection in the airways of cystic fibrosis (8), and susceptibility to salmonella infection in mouse intestinal tracts (9). In addition to In spite of recent advances in the treatment of colon cancer, h multiple liver metastases of colon cancer are still difficult to their direct antimicrobial activities, -defensins are strong treat. Some chemotherapeutic regimens have been reported to chemotactic factors for memory T cells and dendritic cells, be efficient, but there is a high risk of side effects associated suggesting that they also play an important role in acquired immunity (10–12). h-defensins are also inducible by inflammatory with these. Here, we show that isoleucine, an essential amino a h acid, prevents liver metastases in a mouse colon cancer cytokines, such as tumor necrosis factor- and interleukin-1 (13, 14). Recently, Fehlbaum et al. (15) reported that isoleucine metastatic model. Because isoleucine is a strong inducer of h B-defensin, we first hypothesized that it prevented liver and its analogues are highly specific inducers of -defensins. We thus originally hypothesized that isoleucine may contribute to metastases via the accumulation of dendritic cells or memory B tumor immunity through both innate and acquired immunity by T cells through up-regulation of -defensin. However, neither h B-defensin nor immunologic responses were induced by induction of -defensins. -
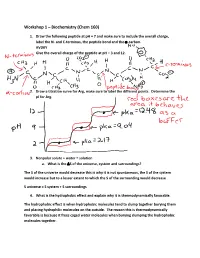
Workshop 1 – Biochemistry (Chem 160)
Workshop 1 – Biochemistry (Chem 160) 1. Draw the following peptide at pH = 7 and make sure to include the overall charge, label the N- and C-terminus, the peptide bond and the -carbon. AVDKY Give the overall charge of the peptide at pH = 3 and 12. 2. Draw a titration curve for Arg, make sure to label the different points. Determine the pI for Arg. 3. Nonpolar solute + water = solution a. What is the S of the universe, system and surroundings? The S of the universe would decrease this is why it is not spontaneous, the S of the system would increase but to a lesser extent to which the S of the surrounding would decrease S universe = S system + S surroundings 4. What is the hydrophobic effect and explain why it is thermodynamically favorable. The hydrophobic effect is when hydrophobic molecules tend to clump together burying them and placing hydrophilic molecules on the outside. The reason this is thermodynamically favorable is because it frees caged water molecules when burying clumping the hydrophobic molecules together. 5. Urea dissolves very readily in water, but the solution becomes very cold as the urea dissolves. How is this possible? Urea dissolves in water because when dissolving there is a net increase in entropy of the universe. The heat exchange, getting colder only reflects the enthalpy (H) component of the total energy change. The entropy change is high enough to offset the enthalpy component and to add up to an overall -G 6. A mutation that changes an alanine residue in the interior of a protein to valine is found to lead to a loss of activity. -

The Optimum Isoleucine:Lysine Ratio in Starter Diets to Maximize Growth Performance of the Early-Weaned Pig
Kansas Agricultural Experiment Station Research Reports Volume 0 Issue 10 Swine Day (1968-2014) Article 837 2000 The optimum isoleucine:lysine ratio in starter diets to maximize growth performance of the early-weaned pig B W. James Robert D. Goodband Michael D. Tokach See next page for additional authors Follow this and additional works at: https://newprairiepress.org/kaesrr Part of the Other Animal Sciences Commons Recommended Citation James, B W.; Goodband, Robert D.; Tokach, Michael D.; Nelssen, Jim L.; and DeRouchey, Joel M. (2000) "The optimum isoleucine:lysine ratio in starter diets to maximize growth performance of the early-weaned pig," Kansas Agricultural Experiment Station Research Reports: Vol. 0: Iss. 10. https://doi.org/10.4148/ 2378-5977.6677 This report is brought to you for free and open access by New Prairie Press. It has been accepted for inclusion in Kansas Agricultural Experiment Station Research Reports by an authorized administrator of New Prairie Press. Copyright 2000 Kansas State University Agricultural Experiment Station and Cooperative Extension Service. Contents of this publication may be freely reproduced for educational purposes. All other rights reserved. Brand names appearing in this publication are for product identification purposes only. No endorsement is intended, nor is criticism implied of similar products not mentioned. K-State Research and Extension is an equal opportunity provider and employer. The optimum isoleucine:lysine ratio in starter diets to maximize growth performance of the early-weaned pig Abstract A total of 360 weanling pigs (initially 12.3 lb BW and approximately 18 d of age) was used in a 14-d growth assay to determine the optimal isoleucine:lysine ratio to maximize growth performance. -

Part One Amino Acids As Building Blocks
Part One Amino Acids as Building Blocks Amino Acids, Peptides and Proteins in Organic Chemistry. Vol.3 – Building Blocks, Catalysis and Coupling Chemistry. Edited by Andrew B. Hughes Copyright Ó 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim ISBN: 978-3-527-32102-5 j3 1 Amino Acid Biosynthesis Emily J. Parker and Andrew J. Pratt 1.1 Introduction The ribosomal synthesis of proteins utilizes a family of 20 a-amino acids that are universally coded by the translation machinery; in addition, two further a-amino acids, selenocysteine and pyrrolysine, are now believed to be incorporated into proteins via ribosomal synthesis in some organisms. More than 300 other amino acid residues have been identified in proteins, but most are of restricted distribution and produced via post-translational modification of the ubiquitous protein amino acids [1]. The ribosomally encoded a-amino acids described here ultimately derive from a-keto acids by a process corresponding to reductive amination. The most important biosynthetic distinction relates to whether appropriate carbon skeletons are pre-existing in basic metabolism or whether they have to be synthesized de novo and this division underpins the structure of this chapter. There are a small number of a-keto acids ubiquitously found in core metabolism, notably pyruvate (and a related 3-phosphoglycerate derivative from glycolysis), together with two components of the tricarboxylic acid cycle (TCA), oxaloacetate and a-ketoglutarate (a-KG). These building blocks ultimately provide the carbon skeletons for unbranched a-amino acids of three, four, and five carbons, respectively. a-Amino acids with shorter (glycine) or longer (lysine and pyrrolysine) straight chains are made by alternative pathways depending on the available raw materials. -

Download Author Version (PDF)
Organic & Biomolecular Chemistry Accepted Manuscript This is an Accepted Manuscript, which has been through the Royal Society of Chemistry peer review process and has been accepted for publication. Accepted Manuscripts are published online shortly after acceptance, before technical editing, formatting and proof reading. Using this free service, authors can make their results available to the community, in citable form, before we publish the edited article. We will replace this Accepted Manuscript with the edited and formatted Advance Article as soon as it is available. You can find more information about Accepted Manuscripts in the Information for Authors. Please note that technical editing may introduce minor changes to the text and/or graphics, which may alter content. The journal’s standard Terms & Conditions and the Ethical guidelines still apply. In no event shall the Royal Society of Chemistry be held responsible for any errors or omissions in this Accepted Manuscript or any consequences arising from the use of any information it contains. www.rsc.org/obc Page 1 of 26 Organic & Biomolecular Chemistry Comparison of alternative nucleophiles for Sortase A-mediated bioconjugation and application in neuronal cell labelling Samuel Baera, Julie Nigro a,b, Mariusz P. Madej a, Rebecca M. Nisbet a,b, Randy Suryadinata a, Gregory Coia a, Lisa P. T. Hong a, Timothy E. Adams a, Charlotte C. Williams *a†, Stewart D. Nuttall a,b†. Manuscript aCSIRO Materials Science and Engineering, 343 Royal Parade, Parkville, Victoria, 3052, AUSTRALIA. bPreventative Health Flagship, 343 Royal Parade, Parkville, Victoria, 3052, AUSTRALIA. *Correspondence to: Charlotte C. Williams ([email protected] ) at CSIRO Materials Accepted Science and Engineering, 343 Royal Parade, Parkville, Victoria, 3052, AUSTRALIA; Ph: +61 3 9662 7100). -

Cysteine Dioxygenase 1 Is a Metabolic Liability for Non-Small Cell Lung Cancer Authors: Yun Pyo Kang1, Laura Torrente1, Min Liu2, John M
bioRxiv preprint doi: https://doi.org/10.1101/459602; this version posted November 1, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Cysteine dioxygenase 1 is a metabolic liability for non-small cell lung cancer Authors: Yun Pyo Kang1, Laura Torrente1, Min Liu2, John M. Asara3,4, Christian C. Dibble5,6 and Gina M. DeNicola1,* Affiliations: 1 Department of Cancer Physiology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL, USA 2 Proteomics and Metabolomics Core Facility, Moffitt Cancer Center and Research Institute, Tampa, FL, USA 3 Division of Signal Transduction, Beth Israel Deaconess Medical Center, Boston, MA, USA 4 Department of Medicine, Harvard Medical School, Boston, MA, USA 5 Department of Pathology and Cancer Center, Beth Israel Deaconess Medical Center, Boston, MA, USA 6 Department of Pathology, Harvard Medical School, Boston, MA, USA *Correspondence to: [email protected]. Keywords: KEAP1, NRF2, cysteine, CDO1, sulfite Summary NRF2 is emerging as a major regulator of cellular metabolism. However, most studies have been performed in cancer cells, where co-occurring mutations and tumor selective pressures complicate the influence of NRF2 on metabolism. Here we use genetically engineered, non-transformed primary cells to isolate the most immediate effects of NRF2 on cellular metabolism. We find that NRF2 promotes the accumulation of intracellular cysteine and engages the cysteine homeostatic control mechanism mediated by cysteine dioxygenase 1 (CDO1), which catalyzes the irreversible metabolism of cysteine to cysteine sulfinic acid (CSA). Notably, CDO1 is preferentially silenced by promoter methylation in non-small cell lung cancers (NSCLC) harboring mutations in KEAP1, the negative regulator of NRF2. -

1 Abietic Acid R Abrasive Silica for Polishing DR Acenaphthene M (LC
1 abietic acid R abrasive silica for polishing DR acenaphthene M (LC) acenaphthene quinone R acenaphthylene R acetal (see 1,1-diethoxyethane) acetaldehyde M (FC) acetaldehyde-d (CH3CDO) R acetaldehyde dimethyl acetal CH acetaldoxime R acetamide M (LC) acetamidinium chloride R acetamidoacrylic acid 2- NB acetamidobenzaldehyde p- R acetamidobenzenesulfonyl chloride 4- R acetamidodeoxythioglucopyranose triacetate 2- -2- -1- -β-D- 3,4,6- AB acetamidomethylthiazole 2- -4- PB acetanilide M (LC) acetazolamide R acetdimethylamide see dimethylacetamide, N,N- acethydrazide R acetic acid M (solv) acetic anhydride M (FC) acetmethylamide see methylacetamide, N- acetoacetamide R acetoacetanilide R acetoacetic acid, lithium salt R acetobromoglucose -α-D- NB acetohydroxamic acid R acetoin R acetol (hydroxyacetone) R acetonaphthalide (α)R acetone M (solv) acetone ,A.R. M (solv) acetone-d6 RM acetone cyanohydrin R acetonedicarboxylic acid ,dimethyl ester R acetonedicarboxylic acid -1,3- R acetone dimethyl acetal see dimethoxypropane 2,2- acetonitrile M (solv) acetonitrile-d3 RM acetonylacetone see hexanedione 2,5- acetonylbenzylhydroxycoumarin (3-(α- -4- R acetophenone M (LC) acetophenone oxime R acetophenone trimethylsilyl enol ether see phenyltrimethylsilyl... acetoxyacetone (oxopropyl acetate 2-) R acetoxybenzoic acid 4- DS acetoxynaphthoic acid 6- -2- R 2 acetylacetaldehyde dimethylacetal R acetylacetone (pentanedione -2,4-) M (C) acetylbenzonitrile p- R acetylbiphenyl 4- see phenylacetophenone, p- acetyl bromide M (FC) acetylbromothiophene 2- -5- -

Amino Acid Chemistry
Handout 4 Amino Acid and Protein Chemistry ANSC 619 PHYSIOLOGICAL CHEMISTRY OF LIVESTOCK SPECIES Amino Acid Chemistry I. Chemistry of amino acids A. General amino acid structure + HN3- 1. All amino acids are carboxylic acids, i.e., they have a –COOH group at the #1 carbon. 2. All amino acids contain an amino group at the #2 carbon (may amino acids have a second amino group). 3. All amino acids are zwitterions – they contain both positive and negative charges at physiological pH. II. Essential and nonessential amino acids A. Nonessential amino acids: can make the carbon skeleton 1. From glycolysis. 2. From the TCA cycle. B. Nonessential if it can be made from an essential amino acid. 1. Amino acid "sparing". 2. May still be essential under some conditions. C. Essential amino acids 1. Branched chain amino acids (isoleucine, leucine and valine) 2. Lysine 3. Methionine 4. Phenyalanine 5. Threonine 6. Tryptophan 1 Handout 4 Amino Acid and Protein Chemistry D. Essential during rapid growth or for optimal health 1. Arginine 2. Histidine E. Nonessential amino acids 1. Alanine (from pyruvate) 2. Aspartate, asparagine (from oxaloacetate) 3. Cysteine (from serine and methionine) 4. Glutamate, glutamine (from α-ketoglutarate) 5. Glycine (from serine) 6. Proline (from glutamate) 7. Serine (from 3-phosphoglycerate) 8. Tyrosine (from phenylalanine) E. Nonessential and not required for protein synthesis 1. Hydroxyproline (made postranslationally from proline) 2. Hydroxylysine (made postranslationally from lysine) III. Acidic, basic, polar, and hydrophobic amino acids A. Acidic amino acids: amino acids that can donate a hydrogen ion (proton) and thereby decrease pH in an aqueous solution 1. -

Figure S1. Heat Map of R (Pearson's Correlation Coefficient)
Figure S1. Heat map of r (Pearson’s correlation coefficient) value among different samples including replicates. The color represented the r value. Figure S2. Distributions of accumulation profiles of lipids, nucleotides, and vitamins detected by widely-targeted UPLC-MC during four fruit developmental stages. The colors indicate the proportional content of each identified metabolites as determined by the average peak response area with R scale normalization. PS1, 2, 3, and 4 represents fruit samples collected at 27, 84, 125, 165 Days After Anthesis (DAA), respectively. Three independent replicates were performed for each stages. Figure S3. Differential metabolites of PS2 vs PS1 group in flavonoid biosynthesis pathway. Figure S4. Differential metabolites of PS2 vs PS1 group in phenylpropanoid biosynthesis pathway. Figure S5. Differential metabolites of PS3 vs PS2 group in flavonoid biosynthesis pathway. Figure S6. Differential metabolites of PS3 vs PS2 group in phenylpropanoid biosynthesis pathway. Figure S7. Differential metabolites of PS4 vs PS3 group in biosynthesis of phenylpropanoids pathway. Figure S8. Differential metabolites of PS2 vs PS1 group in flavonoid biosynthesis pathway and phenylpropanoid biosynthesis pathway combined with RNA-seq results. Table S1. A total of 462 detected metabolites in this study and their peak response areas along the developmental stages of apple fruit. mix0 mix0 mix0 Index Compounds Class PS1a PS1b PS1c PS2a PS2b PS2c PS3a PS3b PS3c PS4a PS4b PS4c ID 1 2 3 Alcohols and 5.25E 7.57E 5.27E 4.24E 5.20E -

Nucleotide Base Coding and Am1ino Acid Replacemients in Proteins* by Emil L
VOL. 48, 1962 BIOCHEMISTRY: E. L. SAIITH 677 18 Britten, R. J., and R. B. Roberts, Science, 131, 32 (1960). '9 Crestfield, A. M., K. C. Smith, and F. WV. Allen, J. Biol. Chem., 216, 185 (1955). 20 Gamow, G., Nature, 173, 318 (1954). 21 Brenner, S., these PROCEEDINGS, 43, 687 (1957). 22 Nirenberg, M. WV., J. H. Matthaei, and 0. WV. Jones, unpublished data. 23 Crick, F. H. C., L. Barnett, S. Brenner, and R. J. Watts-Tobin, Nature, 192, 1227 (1961). 24 Levene, P. A., and R. S. Tipson, J. Biol. Ch-nn., 111, 313 (1935). 25 Gierer, A., and K. W. Mundry, Nature, 182, 1437 (1958). 2' Tsugita, A., and H. Fraenkel-Conrat, J. Mllot. Biol., in press. 27 Tsugita, A., and H. Fraenkel-Conrat, personal communication. 28 Wittmann, H. G., Naturwissenschaften, 48, 729 (1961). 29 Freese, E., in Structure and Function of Genetic Elements, Brookhaven Symposia in Biology, no. 12 (1959), p. 63. NUCLEOTIDE BASE CODING AND AM1INO ACID REPLACEMIENTS IN PROTEINS* BY EMIL L. SMITHt LABORATORY FOR STUDY OF HEREDITARY AND METABOLIC DISORDERS AND THE DEPARTMENTS OF BIOLOGICAL CHEMISTRY AND MEDICINE, UNIVERSITY OF UTAH COLLEGE OF MEDICINE Communicated by Severo Ochoa, February 14, 1962 The problem of which bases of messenger or template RNA' specify the coding of amino acids in proteins has been largely elucidated by the use of synthetic polyri- bonucleotides.2-7 For these triplet nucleotide compositions (Table 1), it is of in- terest to examine some of the presently known cases of amino acid substitutions in polypeptides or proteins of known structure.