Download Author Version (PDF)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Role of Cadaverine and Piperidine in the Formation of N-Nitrosopiperidine in Heated Cured Meat
ROLE OF CADAVERINE AND PIPERIDINE IN THE FORMATION OF N-NITROSOPIPERIDINE IN HEATED CURED MEAT Drabik-Markiewicz G.1, 2, De Mey E. 1, Impens S. 1, Kowalska T. 2, Vander Heyden Y. 3 and Paelinck H.1* 1Research Group for Technology and Quality of Animal Products, Catholic University College Gent, Technology Campus Gent, Department of Chemistry and Biochemistry, 1 Gebroeders Desmetstraat, 9000 Gent, Belgium 2University of Silesia, Institute of Chemistry, 9 Szkolna Street, 40-006 Katowice, Poland 3Analytical Chemistry and Pharmaceutical Technology, Center for Pharmaceutical Research (CePhaR), Vrije Universiteit Brussel (VUB), Laarbeeklaan 103, B-1090 Brussels, Belgium *Corresponding author (e-mail: [email protected]) Abstract — N-nitrosamines are carcinogenic compounds, which formation in meat products depends from different factors e.g., temperature, storage time, precursors and/or added sodium nitrite. Sodium nitrite is important for meat processing as curing agent. The aim of this study was to determine the role of cadaverine and piperidine on the formation of N-nitrosamines in heated cured meat products. Such experimental products were processed with different amounts of sodium nitrite ( 0 mg kg -1, 120 mg kg -1, 480 mg kg -1), 1000 mg kg -1 of cadaverine or 10 mg kg -1 of piperidine, and heated at 85°C, 120°C, 160°C or 220°C. Experimental evidence was produced using gas chromatography in combination with Thermal Energy Analyzer (GC-TEA). The obtained analytical results were statistically evaluated by means of the Univariate Analysis of Variance (ANOVA) approach. In the current study only N-nitrosodimethylamine (NDMA) and N-nitrosopiperidine (NPIP) were detected. -

Article the Bee Hemolymph Metabolome: a Window Into the Impact of Viruses on Bumble Bees
Article The Bee Hemolymph Metabolome: A Window into the Impact of Viruses on Bumble Bees Luoluo Wang 1,2, Lieven Van Meulebroek 3, Lynn Vanhaecke 3, Guy Smagghe 2 and Ivan Meeus 2,* 1 Guangdong Provincial Key Laboratory of Insect Developmental Biology and Applied Technology, Institute of Insect Science and Technology, School of Life Sciences, South China Normal University, Guangzhou, China; [email protected] 2 Department of Plants and Crops, Faculty of Bioscience Engineering, Ghent University, Ghent, Belgium; [email protected], [email protected] 3 Laboratory of Chemical Analysis, Department of Veterinary Public Health and Food Safety, Faculty of Vet- erinary Medicine, Ghent University, Merelbeke, Belgium; [email protected]; [email protected] * Correspondence: [email protected] Selection of the targeted biomarker set: In total we identified 76 metabolites, including 28 amino acids (37%), 11 carbohy- drates (14%), 11 carboxylic acids, 2 TCA intermediates, 4 polyamines, 4 nucleic acids, and 16 compounds from other chemical classes (Table S1). We selected biologically-relevant biomarker candidates based on a three step approach: (1) their expression profile in stand- ardized bees and its relation with viral presence, (2) pathways analysis on significant me- tabolites; and (3) a literature search to identify potential viral specific signatures. Step (1) and (2), pathways analysis on significant metabolites We performed two-way ANOVA with Tukey HSD tests for post-hoc comparisons and used significant metabolites for metabolic pathway analysis using the web-based Citation: Wang, L.L.; Van platform MetaboAnalyst (http://www.metaboanalyst.ca/) in order to get insights in the Meulebroek, L.; Vanhaecke. -

Dietary Supplementation of Inorganic, Organic, and Fatty Acids in Pig: a Review
animals Review Dietary Supplementation of Inorganic, Organic, and Fatty Acids in Pig: A Review Giulia Ferronato * and Aldo Prandini Department of Animal Sciences, Food and Nutrition (DIANA), Faculty of Agriculture, Food and Environmental Science, Università Cattolica del Sacro Cuore, Via Emilia Parmense 84, 29100 Piacenza, Italy; [email protected] * Correspondence: [email protected] Received: 14 August 2020; Accepted: 18 September 2020; Published: 25 September 2020 Simple Summary: The role of acids in pig feed strategies has changed from feed acidifier and preservative to growth promoter and antibiotics substitute. Since the 2006 European banning of growth promoters in the livestock sector, several feed additives have been tested with the goal of identifying molecules with the greatest beneficial antimicrobial, growth-enhancing, or disease-preventing abilities. These properties have been identified among various acids, ranging from inexpensive inorganic acids to organic and fatty acids, and these have been widely used in pig production. Acids are mainly used during the weaning period, which is considered one of the most critical phases in pig farming, as well as during gestation, lactation, and fattening. Such supplementation generally yields improved growth performance and increased feed efficiency; these effects are the consequences of different modes of action acting on the microbiome composition, gut mucosa morphology, enzyme activity, and animal energy metabolism. Abstract: Reduction of antibiotic use has been a hot topic of research over the past decades. The European ban on growth-promoter use has increased the use of feed additivities that can enhance animal growth performance and health status, particularly during critical and stressful phases of life. -

Manipulating Posttranslational Modification in Natural Product Biosynthesis
UC San Diego UC San Diego Electronic Theses and Dissertations Title Manipulating posttranslational modification in natural product biosynthesis Permalink https://escholarship.org/uc/item/0sn8v44w Author Foley, Timothy Leyden Publication Date 2010 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA, SAN DIEGO Manipulating Posttranslational Modification in Natural Product Biosynthesis A dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Chemistry by Timothy Leyden Foley Committee in charge: Professor Michael D. Burkart, Chair Professor Stuart Brody Professor Elizabeth A. Komives Professor Emmanuel Theodorakis Professor Jerry Yang 2010 Copyright Timothy Leyden Foley, 2010 All rights reserved. The dissertation of Timothy Leyden Foley is approved, and it is acceptable in quality and form for publication on microfilm and electronically _____________________________________________________________________ _____________________________________________________________________ _____________________________________________________________________ _____________________________________________________________________ _____________________________________________________________________ Chair University of California, San Diego 2010 iii DEDICATION This dissertation is dedicated to my family and friends. Most of all, to my mother, whose constant support and unwaivering acceptance of my decisions has been -

Phenotype Microarrays™
Phenotype MicroArrays™ PM1 MicroPlate™ Carbon Sources A1 A2 A3 A4 A5 A6 A7 A8 A9 A10 A11 A12 Negative Control L-Arabinose N-Acetyl -D- D-Saccharic Acid Succinic Acid D-Galactose L-Aspartic Acid L-Proline D-Alanine D-Trehalose D-Mannose Dulcitol Glucosamine B1 B2 B3 B4 B5 B6 B7 B8 B9 B10 B11 B12 D-Serine D-Sorbitol Glycerol L-Fucose D-Glucuronic D-Gluconic Acid D,L -α-Glycerol- D-Xylose L-Lactic Acid Formic Acid D-Mannitol L-Glutamic Acid Acid Phosphate C1 C2 C3 C4 C5 C6 C7 C8 C9 C10 C11 C12 D-Glucose-6- D-Galactonic D,L-Malic Acid D-Ribose Tween 20 L-Rhamnose D-Fructose Acetic Acid -D-Glucose Maltose D-Melibiose Thymidine α Phosphate Acid- -Lactone γ D-1 D2 D3 D4 D5 D6 D7 D8 D9 D10 D11 D12 L-Asparagine D-Aspartic Acid D-Glucosaminic 1,2-Propanediol Tween 40 -Keto-Glutaric -Keto-Butyric -Methyl-D- -D-Lactose Lactulose Sucrose Uridine α α α α Acid Acid Acid Galactoside E1 E2 E3 E4 E5 E6 E7 E8 E9 E10 E11 E12 L-Glutamine m-Tartaric Acid D-Glucose-1- D-Fructose-6- Tween 80 -Hydroxy -Hydroxy -Methyl-D- Adonitol Maltotriose 2-Deoxy Adenosine α α ß Phosphate Phosphate Glutaric Acid- Butyric Acid Glucoside Adenosine γ- Lactone F1 F2 F3 F4 F5 F6 F7 F8 F9 F10 F11 F12 Glycyl -L-Aspartic Citric Acid myo-Inositol D-Threonine Fumaric Acid Bromo Succinic Propionic Acid Mucic Acid Glycolic Acid Glyoxylic Acid D-Cellobiose Inosine Acid Acid G1 G2 G3 G4 G5 G6 G7 G8 G9 G10 G11 G12 Glycyl-L- Tricarballylic L-Serine L-Threonine L-Alanine L-Alanyl-Glycine Acetoacetic Acid N-Acetyl- -D- Mono Methyl Methyl Pyruvate D-Malic Acid L-Malic Acid ß Glutamic Acid Acid -

Omega-3 Fatty Acids Correlate with Gut Microbiome Diversity And
Omega-3 fatty acids correlate with gut microbiome diversity and production of N- carbamylglutamate in middle aged and elderly women Cristina Mennia, Jonas Zierera, Tess Pallistera, Matthew A Jacksona, Tao Longb, Robert P Mohneyc, Claire J Stevesa, Tim D Spectora, Ana M Valdesa,d,e a Department of Twin Research and Genetic Epidemiology, Kings College London, London, UK. b Sanford Burnham Prebys, USA cMetabolon Inc., Raleigh-Durham, NC 27709, USA. dSchool of Medicine , Nottingham City Hospital, Hucknall Road, Nottingham, UK. e NIHR Nottingham Biomedical Research Centre, Nottingham, UK. Corresponding author: Dr Ana M Valdes School of Medicine Clinical Sciences Building, Nottingham City Hospital, Hucknall Road, Nottingham, NG5 1PB, UK Phone number: +44 (0)115 823 1954; Fax number:+44(0) 115 823 1757 email: [email protected] Supplementary Table 1. List of faecal metabolites significantly associated to DHA circulating levels (FDR<0.05)adjusting for age, BMI and family relatedness. Super Pathway Sub pathway Metabolite BETA SE P Q Lipid Polyunsaturated Fatty Acid (n3 and n6) eicosapentaenoate (EPA; 20:5n3) 0.15 0.04 4.35x10-5 0.01 Xenobiotics Carbamylated aminoacid N-carbamylglutamate 0.15 0.04 1.21x10-4 0.02 Peptide Dipeptide Derivative anserine 0.13 0.04 5.39x10-4 0.04 Supplementary Table 2. Associations between five measures of microbiome diversity and circulating levels of PUFA adjusting for age, BMI fibre intake and family relatedness SHANNON DIVERSITY CHAO1 OBSERVED Phylogenetic Diversity SIMPSON SPECIES Beta SE P Beta SE P Beta SE P Beta SE P Beta SE P DHA 0.13 0.04 0.002 0.12 0.04 0.003 0.14 0.04 0.001 0.12 0.04 0.01 0.09 0.04 0.02 FAW3 0.12 0.04 0.005 0.13 0.04 0.003 0.13 0.04 0.002 0.12 0.05 0.01 0.08 0.04 0.05 LA 0.10 0.05 0.02 0.10 0.05 0.03 0.10 0.05 0.03 0.10 0.05 0.03 0.09 0.04 0.04 (18:2) FAW6 0.09 0.04 0.02 0.10 0.04 0.01 0.10 0.04 0.01 0.10 0.04 0.02 0.08 0.04 0.05 DHA=22:6 docosahexaenoic acid;, FAW3= Omega-3 fatty acids; 18:2, LA= linoleic acid;; FAW6=omega-6 fatty acids Supplementary Table 3. -

Formation of Cadaverine As an Effect of A-Difluoromethylornithine on Chick Embryo Fibroblasts Transformed by Rous Sarcoma Virus
(CANCER RESEARCH 45, 2159-2164, May 1985] Formation of Cadaverine as an Effect of a-Difluoromethylornithine on Chick Embryo Fibroblasts Transformed by Rous Sarcoma Virus Uriel Bachrach and Alalia Shtorch Department of Molecular Biology, Hebrew University-Hadassah Medical School, Jerusalem, Israel ABSTRACT that DFMO does not interfere with the growth, DNA synthesis, and protein synthesis of normal and transformed cells. However, Chick embryo fibre-blasts grew normally in the presence of 1 DFMO leads to a significant reduction of cellular putrescine x 10~3 to 10 x 1CT3 M a-difluoromethylornithine (DFMO). This levels. This decrease is compensated by the accumulation of drug did not interfere with protein and DNA synthesis of normal cadaverine (1,5-diaminopentane) in the DFMO-treated cells. A fibroblasts and of cells transformed by Rous sarcoma virus. The preliminary account of this work has been given previously (5). morphological appearance of normal and transformed cells was not altered by DFMO, as determined by scanning electron mi croscopy. Flow microfluorometric analyses also confirmed the MATERIALS AND METHODS notion that normal or transformed cells were not blocked by Viruses and Tissue Cultures. A wild type of RSV-SR 17-A and its DFMO in the Gìphase of the cell cycle. As expected, DFMO temperature-sensitive mutant, T5, were used as described elsewhere reduced cellular putrescine levels. This diamine, however, was (10,11). Primary cultures of CEF, prepared from 9- to 11-day-old chick replaced by the analogue cadaverine (1,5-diaminopentane), embryos, were seeded on 90-mm plastic plates (Nunc, Roskilde, Den which accumulated mainly in the transformed cells. -

Table S1. to P 50 and Bo Ttom Loadin G of Primary Com Pone Nt
Table S1. Top 50 and bottom loading of primary component (PC)1 and PC2 among 276 metabolites in the roots of two rice cultivars. Ranking PC1 Loding PC2 Loding Ranking PC1 Loding PC2 Loding 1 β-Ala 0.1133 Mucic acid 0.1344 227 Sebacic acid -0.0017 Pelargonic acid -0.0668 2 Hypotaurine 0.1127 Glucaric acid 0.1247 228 Vanillic acid -0.0020 CoA_divalent -0.0672 3 3-Aminoisobutyric acid 0.1116 Glucosaminic acid 0.1192 229 Trimethylamine -0.0021 Ornithine -0.0676 4 Thr 0.1097 Pyridoxine 0.1180 230 Malic acid -0.0029 Tyr -0.0677 5 Pro 0.1075 Allantoin 0.1158 231 cis-Aconitic acid -0.0030 Arg -0.0680 6 Ser 0.1062 N-Acetylglucosamine 1-phosphate 0.1090 232 Norspermidine -0.0033 γ-Glu-Cys -0.0685 7 O-Acetylserine 0.1051 Trimethylamine 0.1039 233 GMP -0.0038 Adenosine -0.0699 8 cGMP 0.1050 Trimethylamine N-oxide 0.1036 234 2-Hydroxyvaleric acid -0.0044 Citric acid -0.0703 9 Hydroxyproline 0.1032 Betaine 0.1016 235 2-Oxoisovaleric acid -0.0056 Allantoic acid -0.0716 10 N6-Methyllysine 0.1029 Cysteinesulfinic acid 0.1009 236 Uric acid -0.0056 Glucose 6-phosphate -0.0718 11 CMP-N-acetylneuraminate 0.1029 N-Acetyl-β-alanine 0.0991 237 Adenine -0.0071 Rhein -0.0757 12 Glutathione (GSSG)_divalent 0.1020 Pyridoxal 0.0981 238 Adenosine -0.0079 Homoserinelactone -0.0761 13 SDMA 0.1014 Vanillic acid 0.0980 239 Morpholine -0.0082 cis-Aconitic acid -0.0772 14 Phe 0.1014 N-Acetylcysteine 0.0961 240 11-Aminoundecanoic acid -0.0108 Adenine -0.0774 15 5-Aminovaleric acid 0.1014 Phosphoenolpyruvic acid 0.0952 241 1H-Imidazole-4-propionic acid -0.0113 Guanosine -0.0776 -

Induction Kinetics of a Conditional Ph Stress Response System in Escherichia Coli
Induction kinetics of a conditional pH stress response system in Escherichia coli Georg Fritz1, Christiane Koller2, Korinna Burdack2, Larissa Tetsch2, Ina Haneburger2, Kirsten Jung2, and Ulrich Gerland 1;∗ 1 Arnold-Sommerfeld Center for Theoretical Physics (ASC) and Center for NanoScience (CeNS), LMU M¨unchen,Theresienstr. 37, 80333 M¨unchen,Germany 2 Munich Center of integrated Protein Science CiPSM at the Department of Biology I, Microbiology, LMU M¨unchen,Grohaderner Straße 2-4, 82152 Planegg-Martinsried, Germany and ∗Corresponding author, Email address: [email protected] The analysis of stress response systems in microorganisms can reveal molecular strategies for regulatory control and adaptation. Here, we focus on the Cad module, a subsystem of E. coli's response to acidic stress, which is conditionally activated at low pH only when lysine is available. When expressed, the Cad system counteracts the elevated H+concentration by converting lysine to cadaverine under the consumption of H+, and exporting cadaverine in exchange for external lysine. Surprisingly, the cad operon displays a transient response, even when the conditions for its induction persist. To quantitatively characterize the regulation of the Cad module, we have experimentally recorded and theoretically modeled the dynamics of important system variables. We establish a quantitative model that adequately describes and predicts the transient expression behavior for various initial conditions. Our quantitative analysis of the Cad system supports a negative feedback by external cadaverine as the origin of the transient response. Furthermore, the analysis puts causal constraints on the precise mechanism of signal transduction via the regulatory protein CadC. Key words: CadB; CadA; Lysine Decarboxylase; Acid Stress Response; Quantitative Modeling; LysP Introduction cadBA [18]. -

Efficient Production of Enantiopure D-Lysine from L-Lysine by a Two
Article Efficient Production of Enantiopure D‐Lysine from L‐Lysine by a Two‐Enzyme Cascade System Xin Wang, Li Yang, Weijia Cao, Hanxiao Ying, Kequan Chen * and Pingkai Ouyang State Key Laboratory of Materials‐Oriented Chemical Engineering, College of Biotechnology and Pharmaceutical Engineering, Nanjing Tech University, Nanjing 211816, Jiangsu, China; [email protected] (X.W.); [email protected] (L.Y.); [email protected] (W.C.); [email protected] (H.Y.); [email protected] (P.O.) * Correspondence: [email protected]; Tel.: +86‐25‐5813‐9386 Academic Editors: Jose M. Palomo and Cesar Mateo Received: 24 September 2016; Accepted: 25 October 2016; Published: 30 October 2016 Abstract: The microbial production of D‐lysine has been of great interest as a medicinal raw material. Here, a two‐step process for D‐lysine production from L‐lysine by the successive microbial racemization and asymmetric degradation with lysine racemase and decarboxylase was developed. The whole‐cell activities of engineered Escherichia coli expressing racemases from the strains Proteus mirabilis (LYR) and Lactobacillus paracasei (AAR) were first investigated comparatively. When the strain BL21‐LYR with higher racemization activity was employed, L‐lysine was rapidly racemized to give DL‐lysine, and the D‐lysine yield was approximately 48% after 0.5 h. Next, L‐lysine was selectively catabolized to generate cadaverine by lysine decarboxylase. The comparative analysis of the decarboxylation activities of resting whole cells, permeabilized cells, and crude enzyme revealed that the crude enzyme was the best biocatalyst for enantiopure D‐lysine production. The reaction temperature, pH, metal ion additive, and pyridoxal 5′‐phosphate content of this two‐step production process were subsequently optimized. -

Putrescine and Cadaverine in Thehuman
Gut 1993; 34:489-493 489 Presence ofN-acyl and acetoxy derivatives of putrescine and cadaverine in the human gut Gut: first published as 10.1136/gut.34.4.489 on 1 April 1993. Downloaded from K E Murray, K J Shaw, R F Adams, P L Conway Abstract from isolates of gut micro-organisms. Identifica- N-acyl and acetoxy derivatives of putrescine tion of the compounds was obtained in 1-10 .tg and cadaverine have been found in the faeces samples by thin layer chromatography (TLC), of children and in cultures of isolates of gut FDMS, and accurate mass measurement of the bacteria. The evidence was accumulated from molecular ions of the 1-dimethylaminonaphtha- two dimensional, thin layer chromatography, lene-5-sulphonyl- (DANS) derivatives of the field desorption mass spectrometry, and amines concerned. accurate mass measurement of the DANS derivatives of the amines. The acetoxy com- pounds of putrescine and cadaverine have not Methods previously been reported (Gut 1993; 34: 489-493) SUBJECTS AND SOURCES OF SPECIMENS An initial source of faecal specimens was a group of five infants admitted to hospital with salmon- Monoacetylcadaverine (N-acetyl-1,5-diamino- ellosis (Table I). The specimens were taken pentane) and monopropionylcadaverine (N- before antibiotic treatment was begun. A group propionyl-1,5-diaminopentane) have been of 13 children (reported previously6) provided shown to occur frequently in the urine of a second source of faecal specimens. These schizophrenic patients, of psychotic or mentally children had been admitted to hospital for defective patients, and of mentally normal reasons unrelated to gastric problems and were patients in hospital.' These same compounds not receiving any drug treatment. -
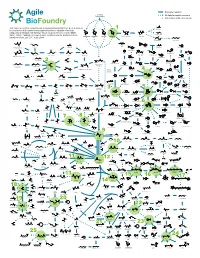
ABF Metabolic Map 11 for Survey Green
Biological reaction Biomass Polysaccharides Multiple biological reactions Potential beachhead molecule HO OH This map represents a compilation of metabolic pathways that have been described Glycolic acid HO HO 1 HO HO for conversion of biomass-derived polysaccharides to industrial chemicals. HO OH O O OH OOH O O O OH O HO OH OH OH OH HO Adapted by permission from Springer Nature Customer Service Centre GmbH: OH OH OH OH OH OH HO OH OH Ethylene glycol Nature, Nature Calalysis, A comprehensive metabolic map for production of bio- OH OH OH OH L-Arabinose Galactose Glucose D-Xylose D-xylono-1,4-lactone Xylonic acid OH OH OH based chemicals, Lee, S.Y., et al., 2019 HO HO 1,2,4-Butanetriol 1,4-Butanediol O OH OH HO OH HO OH O O HO OH HO O OH CoA S OH OH Xylitol HO NH2 HO Propanol Resveratrol Lactaldehyde Malonyl-CoA 4-Hydroxyhexan-3-one HO OH Benzyl alcohol -O Hydroxytyrosol HO O O O CoA S O OH O -O O N HO OH OH O O OH H O 4-phenylbutyrate O OH HO Benzene O O Hippurate Acetol Methylglyoxal Ferulic acid p-Coumaroyl-CoA OH O Propanal 1,2-Propanediol HO 4-Hydroxy-2-oxohexanoic acid p-Hydroxystyrene HO NH2 Benzaldehyde HO OH OH OH Dopamine O OH 1,3-Propanediol O O O O O P - OH O HO O O 2 HO HO HO Glycerone HO OH OH O HO HO 6-phenylhexanoate OH cis, cis-Muconic acid HO O OH O Glyceraldehyde OH HO OH OH HO O O O P - Benzoic acid 3-Phosphate Dihydrobenzenediol Caffeic acid p-Coumaric acid O OH HO 1,3-propanediol 3-Hydroxypropanal HO O HO OH HO NH Glycerol O 2 OH O HO NH2 O HO O P OH Dihydroxyacetone phosphate O OH (R)-Mandelate Pentose L-DOPA phosphate