Boxplots for Grouped and Clustered Data in Toxicology
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
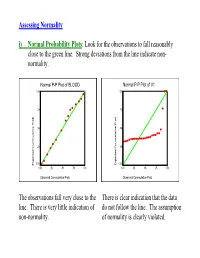
Assessing Normality I) Normal Probability Plots : Look for The
Assessing Normality i) Normal Probability Plots: Look for the observations to fall reasonably close to the green line. Strong deviations from the line indicate non- normality. Normal P-P Plot of BLOOD Normal P-P Plot of V1 1.00 1.00 .75 .75 .50 .50 .25 .25 0.00 0.00 Expected Cummulative Prob Expected Cummulative Expected Cummulative Prob Expected Cummulative 0.00 .25 .50 .75 1.00 0.00 .25 .50 .75 1.00 Observed Cummulative Prob Observed Cummulative Prob The observations fall very close to the There is clear indication that the data line. There is very little indication of do not follow the line. The assumption non-normality. of normality is clearly violated. ii) Histogram: Look for a “bell-shape”. Severe skewness and/or outliers are indications of non-normality. 5 16 14 4 12 10 3 8 2 6 4 1 Std. Dev = 60.29 Std. Dev = 992.11 2 Mean = 274.3 Mean = 573.1 0 N = 20.00 0 N = 25.00 150.0 200.0 250.0 300.0 350.0 0.0 1000.0 2000.0 3000.0 4000.0 175.0 225.0 275.0 325.0 375.0 500.0 1500.0 2500.0 3500.0 4500.0 BLOOD V1 Although the histogram is not perfectly There is a clear indication that the data symmetric and bell-shaped, there is no are right-skewed with some strong clear violation of normality. outliers. The assumption of normality is clearly violated. Caution: Histograms are not useful for small sample sizes as it is difficult to get a clear picture of the distribution. -

Length of Stay Outlier Detection Through Cluster Analysis: a Case Study in Pediatrics Daniel Gartnera, Rema Padmanb
Length of Stay Outlier Detection through Cluster Analysis: A Case Study in Pediatrics Daniel Gartnera, Rema Padmanb aSchool of Mathematics, Cardiff University, United Kingdom bThe H. John Heinz III College, Carnegie Mellon University, Pittsburgh, USA Abstract The increasing availability of detailed inpatient data is enabling the develop- ment of data-driven approaches to provide novel insights for the management of Length of Stay (LOS), an important quality metric in hospitals. This study examines clustering of inpatients using clinical and demographic attributes to identify LOS outliers and investigates the opportunity to reduce their LOS by comparing their order sequences with similar non-outliers in the same cluster. Learning from retrospective data on 353 pediatric inpatients admit- ted for appendectomy, we develop a two-stage procedure that first identifies a typical cluster with LOS outliers. Our second stage analysis compares orders pairwise to determine candidates for switching to make LOS outliers simi- lar to non-outliers. Results indicate that switching orders in homogeneous inpatient sub-populations within the limits of clinical guidelines may be a promising decision support strategy for LOS management. Keywords: Machine Learning; Clustering; Health Care; Length of Stay Email addresses: [email protected] (Daniel Gartner), [email protected] (Rema Padman) 1. Introduction Length of Stay (LOS) is an important quality metric in hospitals that has been studied for decades Kim and Soeken (2005); Tu et al. (1995). However, the increasing digitization of healthcare with Electronic Health Records and other clinical information systems is enabling the collection and analysis of vast amounts of data using advanced data-driven methods that may be par- ticularly valuable for LOS management Gartner (2015); Saria et al. -

Non-Parametric Vs. Parametric Tests in SAS® Venita Depuy and Paul A
NESUG 17 Analysis Perusing, Choosing, and Not Mis-using: ® Non-parametric vs. Parametric Tests in SAS Venita DePuy and Paul A. Pappas, Duke Clinical Research Institute, Durham, NC ABSTRACT spread is less easy to quantify but is often represented by Most commonly used statistical procedures, such as the t- the interquartile range, which is simply the difference test, are based on the assumption of normality. The field between the first and third quartiles. of non-parametric statistics provides equivalent procedures that do not require normality, but often require DETERMINING NORMALITY assumptions such as equal variances. Parametric tests (OR LACK THEREOF) (which assume normality) are often used on non-normal One of the first steps in test selection should be data; even non-parametric tests are used when their investigating the distribution of the data. PROC assumptions are violated. This paper will provide an UNIVARIATE can be implemented to help determine overview of parametric tests and their non-parametric whether or not your data are normal. This procedure equivalents; what assumptions are required for each test; ® generates a variety of summary statistics, such as the how to perform the tests in SAS ; and guidelines for when mean and median, as well as numerical representations of both sets of assumptions are violated. properties such as skewness and kurtosis. Procedures covered will include PROCs ANOVA, CORR, If the population from which the data are obtained is NPAR1WAY, TTEST and UNIVARIATE. Discussion will normal, the mean and median should be equal or close to include assumptions and assumption violations, equal. The skewness coefficient, which is a measure of robustness, and exact versus approximate tests. -

Understanding and Comparing Distributions
Understanding and Comparing Distributions Chapter 4 Objectives: • Boxplot • Calculate Outliers • Comparing Distributions • Timeplot The Big Picture • We can answer much more interesting questions about variables when we compare distributions for different groups. • Below is a histogram of the Average Wind Speed for every day in 1989. The Big Picture (cont.) • The distribution is unimodal and skewed to the right. • The high value may be an outlier Comparing distributions can be much more interesting than just describing a single distribution. The Five-Number Summary • The five-number summary of a distribution reports its Max 8.67 median, quartiles, and Q3 2.93 extremes (maximum and minimum). Median 1.90 • Example: The five-number Q1 1.15 summary for for the daily Min 0.20 wind speed is: The Five-Number Summary • Consists of the minimum value, Q1, the median, Q3, and the maximum value, listed in that order. • Offers a reasonably complete description of the center and spread. • Calculate on the TI-83/84 using 1-Var Stats. • Used to construct the Boxplot. • Example: Five-Number Summary • 1: 20, 27, 34, 50, 86 • 2: 5, 10, 18.5, 29, 33 Daily Wind Speed: Making Boxplots • A boxplot is a graphical display of the five- number summary. • Boxplots are useful when comparing groups. • Boxplots are particularly good at pointing out outliers. Boxplot • A graph of the Five-Number Summary. • Can be drawn either horizontally or vertically. • The box represents the IQR (middle 50%) of the data. • Show less detail than histograms or stemplots, they are best used for side-by-side comparison of more than one distribution. -

Notes Unit 8: Interquartile Range, Box Plots, and Outliers I
Notes Unit 8: Interquartile Range, Box Plots, and Outliers I. Box Plot A. What is it? • Also called a ‘Box and Whiskers’ plot • A 5-numbered summary of data: • Lower extreme • Lower quartile • Median • Upper quartile • Upper extreme • To draw a Box Plot, we need to find all 5 of these numbers B. Steps to Creating a Box Plot 1. Order the numbers smallest to largest 2. Find the 5 numbers- median, lower and upper extremes, lower and upper quartiles 3 Draw the box plot- draw a number line, draw and label the parts C. Examples Example 1: 12, 13, 5, 8, 9, 20, 16, 14, 14, 6, 9, 12, 12 Step 1: Order the numbers from smallest to largest 5, 6, 8, 9, 9, 12, 12, 12, 13, 14, 14, 16, 20 Step 2 – Find the Median 5, 6, 8, 9, 9, 12, 12, 12, 13, 14, 14, 16, 20 Median: 12 2. Find the median. The median is the middle number. If the data has two middle numbers, find the mean of the two numbers. What is the median? Step 2 – Find the Lower and Upper Extremes 5, 6, 8, 9, 9, 12, 12, 12, 13, 14, 14, 16, 20 Median: 12 5 20 2. Find the smallest and largest numbers Step 2 – Upper & Lower Quartiles 5, 6, 8, 9, 9, 12, 12, 12, 13, 14, 14, 16, 20 Median: 5 lower 12 upper 20 quartile: quartile: 8.5 14 3. Find the lower and upper medians or quartiles. These are the middle numbers on each side of the median. -

5. Drawing Graphs
5. Drawing graphs Above all else show the data. –Edward Tufte1 Visualising data is one of the most important tasks facing the data analyst. It’s important for two distinct but closely related reasons. Firstly, there’s the matter of drawing “presentation graphics”, displaying your data in a clean, visually appealing fashion makes it easier for your reader to understand what you’re trying to tell them. Equally important, perhaps even more important, is the fact that drawing graphs helps you to understand the data. To that end, it’s important to draw “exploratory graphics” that help you learn about the data as you go about analysing it. These points might seem pretty obvious but I cannot count the number of times I’ve seen people forget them. To give a sense of the importance of this chapter, I want to start with a classic illustration of just how powerful a good graph can be. To that end, Figure 5.1 shows a redrawing of one of the most famous data visualisations of all time. This is John Snow’s 1854 map of cholera deaths. The map is elegant in its simplicity. In the background we have a street map which helps orient the viewer. Over the top we see a large number of small dots, each one representing the location of a cholera case. The larger symbols show the location of water pumps, labelled by name. Even the most casual inspection of the graph makes it very clear that the source of the outbreak is almost certainly the Broad Street pump. -

A Simple Glossary of Statistics. Barchart: Box-Plot: (Also Known As
1 A simple glossary of statistics. Barchart: 3.5 Similar to a Histogram but the bars don‟t touch each other and the x-axis usually does not have a 3.0 continuous scale. 2.5 The example shows a bar chart of the colour of car 2.0 owned by ten people. (Note how the false origin 1.5 gives a first impression of even less blue cars.) 1.0 Count .5 red green black w hite blue Box-plot: (also known as box CARCOLOR and whisker plot) A Boxplot divides the data into quarters. The middle line shows the median (the value that Boxplot divides the data in half), the box shows the 110 range of the two middle quarters, and the 100 whisker whiskers show the range of the rest of the data. 90 upper quartile The values at the ends of the box are called the t h th g i 80 e quartiles, (SPSS refers to these as the 25 and W box th 70 m edian 75 percentiles) The distance between them is lower quartile 60 called the interquartile range (IQR). whisker 50 The more sophisticated version (which SPSS uses) marks outliers with circles, counting anything more than one and a half times the interquartile range away from the quartiles as an outlier, those over three times the interquartile range away from the quartiles are called extremes and marked with asterisks. The length of the box is equal to the interquartile range (IQR). Boxplots are most often used for comparing two or more sets of data. -

Kruskal- Wallis Test Is the Non-Parametric Equivalent to 0.95 1.76 2.91 One-Way ANOVA
community project encouraging academics to share statistics support resources All stcp resources are released under a Creative Commons licence stcp-marshall-KruskalSPSS The following resources are associated: One Way ANOVA quick reference worksheet and Mann-Whitney U Test worksheet Kruskal-Wallis Test in SPSS (Non-parametric equivalent to ANOVA) Research question type: Differences between several groups of measurements Dependent variable: Continuous (scale) but not normally distributed or ordinal Independent variable: Categorical (Nominal) Common Applications: Comparing the mean rank of three or more different groups in scientific or medical experiments when the dependent variable ordinal or is not normally distributed. Descriptive statistics: Median for each group and box-plot Example: Alcohol, coffee and reaction times Reaction time An experiment was carried out to see if alcohol or coffee Water Coffee Alcohol affects driving reaction times. There were three different 0.37 0.98 1.69 groups of participants; 10 drinking water, 10 drinking beer containing two units of alcohol and 10 drinking coffee. 0.38 1.11 1.71 Reaction time on a driving simulation was measured for 0.61 1.27 1.75 each participant. 0.78 1.32 1.83 0.83 1.44 1.97 The sample sizes are small and normality checks showed that the assumption of normality had not been met so an 0.86 1.45 2.53 ANOVA is not suitable for comparing the groups. The 0.9 1.46 2.66 Kruskal- Wallis test is the non-parametric equivalent to 0.95 1.76 2.91 one-way ANOVA. 1.63 2.56 3.28 1.97 3.07 3.47 © Ellen Marshall Reviewer: Peter Samuels Sheffield Hallam University Birmingham City University Kruskal-Wallis in SPSS The median scores show that reaction time after coffee and alcohol is higher than for those drinking just water. -

Measures of Central Tendency and Spread & Box-And-Whisker Plots
Algebra 1 Name_______________________________ Notes – 8.1 Measures of Central Tendency and Spread & Box-and-Whisker Plots Opener – Find the average of the given set of data. 23 25 34 36 19 25 17 23 22 39 36 Measures of Central Tendency mean – median – mode – Example 1: Find the measures of central tendencies given the set of data. The tuition costs (in thousands of dollars) for ten different liberal arts colleges are listed below. 38 33 40 27 44 32 23 27 47 31 Think About It! A – If the data value of “23” were changed to “26”, what would be the effect on the mean, median, and mode? B – If the data value of “33” were changed to “32”, what would be the effect on the mean, median, and mode? Practice 2: Find the measure of central tendencies given the set of data. The tuition costs (in thousands of dollars) for ten different liberal arts colleges are listed below. 43 33 31 40 31 27 42 47 32 41 The Five Number Summary min = Q1 = med = Q3 = max = Measures of Spread range – lower quartile range – upper quartile range – interquartile range (IQR) – Example 3: Use the data given to find the following. 22 17 18 29 22 22 23 24 23 17 21 Minimum = Q1 = Median = Q3 = Maximum = Range = IQR = Box-and-Whisker Plot Example 4: The last 11 credit card purchases (rounded to the nearest dollar) for someone are given. 22 66 44 91 12 45 48 31 20 82 19 Fill in each value and the create a box-and-whisker plot. -

Trajectory Box Plot: a New Pattern to Summarize Movements Laurent Etienne, Thomas Devogele, Maike Buckin, Gavin Mcardle
Trajectory Box Plot: a new pattern to summarize movements Laurent Etienne, Thomas Devogele, Maike Buckin, Gavin Mcardle To cite this version: Laurent Etienne, Thomas Devogele, Maike Buckin, Gavin Mcardle. Trajectory Box Plot: a new pattern to summarize movements. International Journal of Geographical Information Science, Taylor & Francis, 2016, Analysis of Movement Data, 30 (5), pp.835-853. 10.1080/13658816.2015.1081205. hal-01215945 HAL Id: hal-01215945 https://hal.archives-ouvertes.fr/hal-01215945 Submitted on 25 Apr 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. August 4, 2015 10:29 International Journal of Geographical Information Science trajec- tory_boxplot_ijgis_20150722 International Journal of Geographical Information Science Vol. 00, No. 00, January 2015, 1{20 RESEARCH ARTICLE Trajectory Box Plot; a New Pattern to Summarize Movements Laurent Etiennea∗, Thomas Devogelea, Maike Buchinb, Gavin McArdlec aUniversity of Tours, Blois, France; bFaculty of Mathematics, Ruhr University, Bochum, Deutschland; cNational Center for Geocomputation, Maynooth University, Ireland; (v2.1 sent July 2015) Nowadays, an abundance of sensors are used to collect very large datasets of moving objects. The movement of these objects can be analysed by identify- ing common routes. -

Constructing Box Plots
CONSTRUCTING BOX PLOTS 6.12A The student is expected to represent numeric data graphically, including dot plots, stem-and-leaf plots, histograms, and box plots. TELL ME MORE… A graph helps you visualize certain characteristics of a data set. Some graphs, like dot plots, stem-and-leaf plots, or histograms, help you see the distribution of the data within the set. In other words, you can see characteristics such as which data values are more common than others or how close in value the data values are. Other types of graphs, such as box plots (also called box-and-whiskers plots) show you how spread out the data values in the set are. Box plots have a box that shows where the middle half of the data set is. Each end has a whisker that shows you the lower fourth and upper fourth of the data set. The football team at Ann Richards Middle School played 11 games last season. The number of points the team scored in each game is shown in the following list. 12, 28, 40, 6, 24, 16, 20, 21, 14, 22, 36 You can represent the data set with a box plot. First, you will need to determine the median, frst quartile, and third quartile. To help with calculating these values, list the data set in order from least to greatest. 6, 12, 14, 16, 20, 21, 22, 24, 28, 36, 40 The first quartile is the The third quartile is the middle value of the frst middle value of the second half of the data (all of the half of the data (all of the numbers less than the The median is the middle numbers greater than the median). -

Boxplots, Interquartile Range, and Outliers
Boxplots, Interquartile Range, and Outliers Boxplots provide a visual representation of a data set that can be used to determine whether the data set is symmetric or skewed. Constructing a boxplot requires calculation of the “5 number summary”, the interquartile range (IQR), and the presence of any outliers. 5 Number Summary – The 5 number summary for a data set includes the following, which are listed in order from smallest to largest – 1. Minimum - The smallest value in the data set. 2. First Quartile - Separates the lowest 25% of the data in a set from the highest 75%. It is 25 typically denoted as 푸 푤ℎ푒푟푒, ∙ (# 푝표푛푡푠 푛 푑푎푡푎 푠푒푡) = 푝표푠푡표푛 표푓 푄 푛 푠푒푡. ퟏ 100 1 3. Median – The middle value in a sorted (smallest to largest) data set. If there is an even number of values, it is calculated by averaging the two middle values. The Median is also referred to as the Second Quartile (푸ퟐ) because it separates the lower 50% of data in a set from the upper 50%. 4. Third Quartile - Separates the lowest 75% of the data in a set from the highest 25%. It is 75 typically denoted as 푸 푤ℎ푒푟푒, ∙ (# 푝표푛푡푠 푛 푑푎푡푎 푠푒푡) = 푝표푠푡표푛 표푓 푄 푛 푠푒푡. ퟑ 100 3 5. Maximum – The largest value in the data set. IQR - The Interquartile Range is a measure of spread used to calculate the lower and upper outlier boundaries. These boundaries are then used to determine whether a data set has any actual outliers. 푰풏풕풆풓풒풖풂풓풕풊풍풆 푹풂풏품풆 (퐼푄푅) = 푄3 − 푄1 푳풐풘풆풓 푂푢푡푙푒푟 퐵표푢푛푑푎푟푦 = 푄1 − 1.5 퐼푄푅 푼풑풑풆풓 푂푢푡푙푒푟 퐵표푢푛푑푎푟푦 = 푄3 + 1.5 퐼푄푅 Outliers - Outliers are data points that are considerably smaller or larger than most of the other values in a data set.