5. Drawing Graphs
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
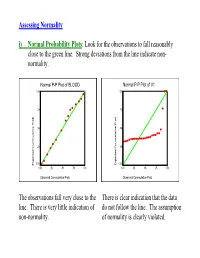
Assessing Normality I) Normal Probability Plots : Look for The
Assessing Normality i) Normal Probability Plots: Look for the observations to fall reasonably close to the green line. Strong deviations from the line indicate non- normality. Normal P-P Plot of BLOOD Normal P-P Plot of V1 1.00 1.00 .75 .75 .50 .50 .25 .25 0.00 0.00 Expected Cummulative Prob Expected Cummulative Expected Cummulative Prob Expected Cummulative 0.00 .25 .50 .75 1.00 0.00 .25 .50 .75 1.00 Observed Cummulative Prob Observed Cummulative Prob The observations fall very close to the There is clear indication that the data line. There is very little indication of do not follow the line. The assumption non-normality. of normality is clearly violated. ii) Histogram: Look for a “bell-shape”. Severe skewness and/or outliers are indications of non-normality. 5 16 14 4 12 10 3 8 2 6 4 1 Std. Dev = 60.29 Std. Dev = 992.11 2 Mean = 274.3 Mean = 573.1 0 N = 20.00 0 N = 25.00 150.0 200.0 250.0 300.0 350.0 0.0 1000.0 2000.0 3000.0 4000.0 175.0 225.0 275.0 325.0 375.0 500.0 1500.0 2500.0 3500.0 4500.0 BLOOD V1 Although the histogram is not perfectly There is a clear indication that the data symmetric and bell-shaped, there is no are right-skewed with some strong clear violation of normality. outliers. The assumption of normality is clearly violated. Caution: Histograms are not useful for small sample sizes as it is difficult to get a clear picture of the distribution. -

If I Were Doing a Quick ANOVA Analysis
ANOVA Note: If I were doing a quick ANOVA analysis (without the diagnostic checks, etc.), I’d do the following: 1) load the packages (#1); 2) do the prep work (#2); and 3) run the ggstatsplot::ggbetweenstats analysis (in the #6 section). 1. Packages needed. Here are the recommended packages to load prior to working. library(ggplot2) # for graphing library(ggstatsplot) # for graphing and statistical analyses (one-stop shop) library(GGally) # This package offers the ggpairs() function. library(moments) # This package allows for skewness and kurtosis functions library(Rmisc) # Package for calculating stats and bar graphs of means library(ggpubr) # normality related commands 2. Prep Work Declare factor variables as such. class(mydata$catvar) # this tells you how R currently sees the variable (e.g., double, factor) mydata$catvar <- factor(mydata$catvar) #Will declare the specified variable as a factor variable 3. Checking data for violations of assumptions: a) relatively equal group sizes; b) equal variances; and c) normal distribution. a. Group Sizes Group counts (to check group frequencies): table(mydata$catvar) b. Checking Equal Variances Group means and standard deviations (Note: the aggregate and by commands give you the same results): aggregate(mydata$intvar, by = list(mydata$catvar), FUN = mean, na.rm = TRUE) aggregate(mydata$intvar, by = list(mydata$catvar), FUN = sd, na.rm = TRUE) by(mydata$intvar, mydata$catvar, mean, na.rm = TRUE) by(mydata$intvar, mydata$catvar, sd, na.rm = TRUE) A simple bar graph of group means and CIs (using Rmisc package). This command is repeated further below in the graphing section. The ggplot command will vary depending on the number of categories in the grouping variable. -

Healthy Volunteers (Retrospective Study) Urolithiasis Healthy Volunteers Group P-Value 5 (N = 110) (N = 157)
Supplementary Table 1. The result of Gal3C-S-OPN between urolithiasis and healthy volunteers (retrospective study) Urolithiasis Healthy volunteers Group p-Value 5 (n = 110) (n = 157) median (IQR 1) median (IQR 1) Gal3C-S-OPN 2 515 1118 (810–1543) <0.001 (MFI 3) (292–786) uFL-OPN 4 14 56392 <0.001 (ng/mL/mg protein) (10–151) (30270-115516) Gal3C-S-OPN 2 52 0.007 /uFL-OPN 4 <0.001 (5.2–113.0) (0.003–0.020) (MFI 3/uFL-OPN 4) 1 IQR, Interquartile range; 2 Gal3C-S-OPN, Gal3C-S lectin reactive osteopontin; 3 MFI, mean fluorescence intensity; 4 uFL-OPN, Urinary full-length-osteopontin; 5 p-Value, Mann–Whitney U-test. Supplementary Table 2. Sex-related difference between Gal3C-S-OPN and Gal3C-S-OPN normalized to uFL- OPN (retrospective study) Group Urolithiasis Healthy volunteers p-Value 5 Male a Female b Male c Female d a vs. b c vs. d (n = 61) (n = 49) (n = 57) (n = 100) median (IQR 1) median (IQR 1) Gal3C-S-OPN 2 1216 972 518 516 0.15 0.28 (MFI 3) (888-1581) (604-1529) (301-854) (278-781) Gal3C-S-OPN 2 67 42 0.012 0.006 /uFL-OPN 4 0.62 0.56 (9-120) (4-103) (0.003-0.042) (0.002-0.014) (MFI 3/uFL-OPN 4) 1 IQR, Interquartile range; 2 Gal3C-S-OPN, Gal3C-S lectin reactive osteopontin; 3MFI, mean fluorescence intensity; 4 uFL-OPN, Urinary full-length-osteopontin; 5 p-Value, Mann–Whitney U-test. -

Violin Plots: a Box Plot-Density Trace Synergism Author(S): Jerry L
Violin Plots: A Box Plot-Density Trace Synergism Author(s): Jerry L. Hintze and Ray D. Nelson Source: The American Statistician, Vol. 52, No. 2 (May, 1998), pp. 181-184 Published by: American Statistical Association Stable URL: http://www.jstor.org/stable/2685478 Accessed: 02/09/2010 11:01 Your use of the JSTOR archive indicates your acceptance of JSTOR's Terms and Conditions of Use, available at http://www.jstor.org/page/info/about/policies/terms.jsp. JSTOR's Terms and Conditions of Use provides, in part, that unless you have obtained prior permission, you may not download an entire issue of a journal or multiple copies of articles, and you may use content in the JSTOR archive only for your personal, non-commercial use. Please contact the publisher regarding any further use of this work. Publisher contact information may be obtained at http://www.jstor.org/action/showPublisher?publisherCode=astata. Each copy of any part of a JSTOR transmission must contain the same copyright notice that appears on the screen or printed page of such transmission. JSTOR is a not-for-profit service that helps scholars, researchers, and students discover, use, and build upon a wide range of content in a trusted digital archive. We use information technology and tools to increase productivity and facilitate new forms of scholarship. For more information about JSTOR, please contact [email protected]. American Statistical Association is collaborating with JSTOR to digitize, preserve and extend access to The American Statistician. http://www.jstor.org StatisticalComputing and Graphics ViolinPlots: A Box Plot-DensityTrace Synergism JerryL. -

Statistical Analysis in JASP
Copyright © 2018 by Mark A Goss-Sampson. All rights reserved. This book or any portion thereof may not be reproduced or used in any manner whatsoever without the express written permission of the author except for the purposes of research, education or private study. CONTENTS PREFACE .................................................................................................................................................. 1 USING THE JASP INTERFACE .................................................................................................................... 2 DESCRIPTIVE STATISTICS ......................................................................................................................... 8 EXPLORING DATA INTEGRITY ................................................................................................................ 15 ONE SAMPLE T-TEST ............................................................................................................................. 22 BINOMIAL TEST ..................................................................................................................................... 25 MULTINOMIAL TEST .............................................................................................................................. 28 CHI-SQUARE ‘GOODNESS-OF-FIT’ TEST............................................................................................. 30 MULTINOMIAL AND Χ2 ‘GOODNESS-OF-FIT’ TEST. .......................................................................... -

Beanplot: a Boxplot Alternative for Visual Comparison of Distributions
Beanplot: A Boxplot Alternative for Visual Comparison of Distributions Peter Kampstra VU University Amsterdam Abstract This introduction to the R package beanplot is a (slightly) modified version of Kamp- stra(2008), published in the Journal of Statistical Software. Boxplots and variants thereof are frequently used to compare univariate data. Boxplots have the disadvantage that they are not easy to explain to non-mathematicians, and that some information is not visible. A beanplot is an alternative to the boxplot for visual comparison of univariate data between groups. In a beanplot, the individual observations are shown as small lines in a one-dimensional scatter plot. Next to that, the estimated density of the distributions is visible and the average is shown. It is easy to compare different groups of data in a beanplot and to see if a group contains enough observations to make the group interesting from a statistical point of view. Anomalies in the data, such as bimodal distributions and duplicate measurements, are easily spotted in a beanplot. For groups with two subgroups (e.g., male and female), there is a special asymmetric beanplot. For easy usage, an implementation was made in R. Keywords: exploratory data analysis, descriptive statistics, box plot, boxplot, violin plot, density plot, comparing univariate data, visualization, beanplot, R, graphical methods, visu- alization. 1. Introduction There are many known plots that are used to show distributions of univariate data. There are histograms, stem-and-leaf-plots, boxplots, density traces, and many more. Most of these plots are not handy when comparing multiple batches of univariate data. For example, com- paring multiple histograms or stem-and-leaf plots is difficult because of the space they take. -

Boxplots for Grouped and Clustered Data in Toxicology
Penultimate version. If citing, please refer instead to the published version in Archives of Toxicology, DOI: 10.1007/s00204-015-1608-4. Boxplots for grouped and clustered data in toxicology Philip Pallmann1, Ludwig A. Hothorn2 1 Department of Mathematics and Statistics, Lancaster University, Lancaster LA1 4YF, UK, Tel.: +44 (0)1524 592318, [email protected] 2 Institute of Biostatistics, Leibniz University Hannover, 30419 Hannover, Germany Abstract The vast majority of toxicological papers summarize experimental data as bar charts of means with error bars. While these graphics are easy to generate, they often obscure essen- tial features of the data, such as outliers or subgroups of individuals reacting differently to a treatment. Especially raw values are of prime importance in toxicology, therefore we argue they should not be hidden in messy supplementary tables but rather unveiled in neat graphics in the results section. We propose jittered boxplots as a very compact yet comprehensive and intuitively accessible way of visualizing grouped and clustered data from toxicological studies together with individual raw values and indications of statistical significance. A web application to create these plots is available online. Graphics, statistics, R software, body weight, micronucleus assay 1 Introduction Preparing a graphical summary is usually the first if not the most important step in a data analysis procedure, and it can be challenging especially with many-faceted datasets as they occur frequently in toxicological studies. However, even in simple experimental setups many researchers have a hard time presenting their results in a suitable manner. Browsing recent volumes of this journal, we have realized that the least favorable ways of displaying toxicological data appear to be the most popular ones (according to the number of publications that use them). -

Length of Stay Outlier Detection Through Cluster Analysis: a Case Study in Pediatrics Daniel Gartnera, Rema Padmanb
Length of Stay Outlier Detection through Cluster Analysis: A Case Study in Pediatrics Daniel Gartnera, Rema Padmanb aSchool of Mathematics, Cardiff University, United Kingdom bThe H. John Heinz III College, Carnegie Mellon University, Pittsburgh, USA Abstract The increasing availability of detailed inpatient data is enabling the develop- ment of data-driven approaches to provide novel insights for the management of Length of Stay (LOS), an important quality metric in hospitals. This study examines clustering of inpatients using clinical and demographic attributes to identify LOS outliers and investigates the opportunity to reduce their LOS by comparing their order sequences with similar non-outliers in the same cluster. Learning from retrospective data on 353 pediatric inpatients admit- ted for appendectomy, we develop a two-stage procedure that first identifies a typical cluster with LOS outliers. Our second stage analysis compares orders pairwise to determine candidates for switching to make LOS outliers simi- lar to non-outliers. Results indicate that switching orders in homogeneous inpatient sub-populations within the limits of clinical guidelines may be a promising decision support strategy for LOS management. Keywords: Machine Learning; Clustering; Health Care; Length of Stay Email addresses: [email protected] (Daniel Gartner), [email protected] (Rema Padman) 1. Introduction Length of Stay (LOS) is an important quality metric in hospitals that has been studied for decades Kim and Soeken (2005); Tu et al. (1995). However, the increasing digitization of healthcare with Electronic Health Records and other clinical information systems is enabling the collection and analysis of vast amounts of data using advanced data-driven methods that may be par- ticularly valuable for LOS management Gartner (2015); Saria et al. -

Chapter 6: Graphics
Chapter 6 Graphics Modern computers with high resolution displays and graphic printers have rev- olutionized the visual display of information in fields ranging from computer- aided design, through flow dynamics, to the spatiotemporal attributes of infec- tious diseases. The impact on statistics is just being felt. Whole books have been written on statistical graphics and their contents are quite heterogeneous– simple how-to-do it texts (e.g., ?; ?), reference works (e.g., Murrell, 2006) and generic treatment of the principles of graphic presentation (e.g., ?). There is even a web site devoted to learning statistics through visualization http: //www.seeingstatistics.com/.Hence,itisnotpossibletobecomprehensive in this chapter. Instead, I focus on the types of graphics used most often in neu- roscience (e.g., plots of means) and avoid those seldom used in the field (e.g., pie charts, geographical maps). The here are four major purposes for statistical graphics. First, they are used to examine and screen data to check for abnormalities and to assess the distributions of the variables. Second, graphics are very useful aid to exploratory data analysis (??). Exploratory data analysis, however, is used for mining large data sets mostly for the purpose of hypothesis generation and modification, so that use of graphics will not be discussed here. Third, graphics can be used to assess both the assumptions and the validity of a statistical model applied to data. Finally, graphics are used to present data to others. The third of these purposes will be discussed in the appropriate sections on the statistics. This chapter deals with the first and last purposes–examining data and presenting results. -

Dynamics of Hydrological Model Parameters: Calibration and Reliability
Dynamics of hydrological model parameters: calibration and reliability Tian Lan1, Kairong Lin1,2,3, Xuezhi Tan1,2,3, Chong-Yu Xu4, Xiaohong Chen1,2,3 1Center for Water Resources and Environment, Sun Yat-sen University, Guangzhou, 510275, China. 2Guangdong Engineering Technology Research Center of Water Security Regulation and Control for Southern China, Guangzhou 510275, China. 3School of Civil Engineering, Sun Yat-sen University, Guangzhou, 510275, China. 4Department of Geosciences, University of Oslo, P.O. Box 1047, Blindern, 0316 Oslo, Norway Correspondence to: Kairong Lin ([email protected]) Contents of this file 1 Case study and data 2 HYMOD model 3 SCE-UA 4 Violin plot 5 Multi-metric evaluation 6 Evaluation of sub-period calibration schemes in Mumahe basin and Xunhe basin 7 Convergence performance in Mumahe basin and Xunhe basin using ECP-VP References Introduction This supporting information includes eight sections that support the analysis. The 1 Case study and data section is used to support the 2 Background section in the main manuscript. The 2 HYMOD model section is used to support the 3.1 Sub-period calibration section in the main manuscript. The 3 SCE-UA algorithm section and 4 Violin plot section are used to support the 3.2 A tool for reliability evaluation section in the main manuscript. The 5 Multi-metric evaluation section and 6 Evaluation of sub-period calibration schemes in Mumahe basin and Xunhe basin section are used to account for 4.1 Evaluation of calibration schemes section in the main manuscript. The 7 Convergence performance in Mumahe basin and Xunhe basin using ECP-VP section is used to supplement 4.3 Evaluation of reliability section in the main manuscript. -

Modern Statistics for Modern Biology
Contents 3 High Quality Graphics in R 5 3.1 Goals for this Chapter .......................... 5 3.2 Base R Plotting .............................. 6 3.3 An Example Dataset ........................... 7 3.4 ggplot2 .................................. 8 3.5 The Grammar of Graphics ........................ 9 3.6 1D Visualisations ............................. 12 3.6.1 Data tidying I – matrices versus data.frame . 12 3.6.2 Barplots ............................. 13 3.6.3 Boxplots ............................. 13 3.6.4 Violin plots ........................... 13 3.6.5 Dot plots and beeswarm plots . 14 3.6.6 Density plots ........................... 14 3.6.7 ECDF plots ............................ 15 3.6.8 Transformations ......................... 15 3.6.9 Data tidying II - wide vs long format . 16 3.7 2D Visualisation: Scatter Plots ...................... 17 3.7.1 Plot shapes ............................ 18 3.8 3–5D Data ................................. 19 3.8.1 Faceting ............................. 21 3.8.2 Interactive graphics ....................... 22 3.9 Colour ................................... 23 3.9.1 Heatmaps ............................ 24 3.9.2 Colour spaces .......................... 26 3.10 Data Transformations .......................... 27 3.11 Saving Figures .............................. 27 3.12 Biological Data with ggbio ........................ 28 3.13 Recap of this Chapter ........................... 29 3.14 Exercises ................................. 29 6 Multiple Testing 31 6.1 Goals for this Chapter ......................... -

Moving Beyond P Values: Everyday Data Analysis with Estimation Plots
bioRxiv preprint doi: https://doi.org/10.1101/377978 ; this version posted April 6, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Moving beyond P values: Everyday data analysis with estimation plots Joses Ho 1, Tayfun Tumkaya 1, 2, Sameer Aryal 1, 3 , Hyungwon Choi 1, 4, Adam Claridge-Chang 1, 2, 5, 6 1. Institute for Molecular and Cell Biology, A*STAR, Singapore 138673 2. Department of Physiology, National University of Singapore, Singapore 3. Center for Neural Science, New York University, New York, NY, USA 4. Department of Medicine, Yong Loo Lin School of Medicine, National University of Singapore, Singapore 5. Program in Neuroscience and Behavioral Disorders, Duke-NUS Medical School, Singapore 6. Correspondence Introduction Over the past 75 years, a number of statisticians have advised that the data-analysis method known as null-hypothesis significance testing (NHST) should be deprecated (Berkson, 1942; Halsey et al., 2015; Wasserstein et al., 2019). The limitations of NHST have been extensively discussed, with a broad consensus that current statistical practice in the biological sciences needs reform. However, there is less agreement on reform’s specific nature, with vigorous debate surrounding what would constitute a suitable alternative (Altman et al., 2000; Benjamin et al., 2017; Cumming and Calin-Jageman, 2016). An emerging view is that a more complete analytic technique would use statistical graphics to estimate effect sizes and evaluate their uncertainty (Cohen, 1994; Cumming and Calin-Jageman, 2016).