Rap1 Gtpase: Functions, Regulation, and Malignancy
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
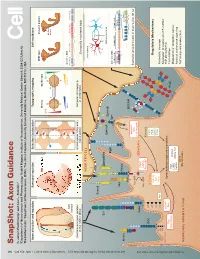
Snapshot: Axon Guidance Pasterkamp R
494 1 Cell Cell ??? SnapShot: Axon Guidance 153 SnapShot: XXXXXXXXXXXXXXXXXXXXXXXXXX 1 2 , ??MONTH?? ??DATE??, 200? ©200? Elsevier Inc. 200?©200? ElsevierInc. , ??MONTH?? ??DATE??, DOI R. Jeroen Pasterkamp and Alex L. Kolodkin , April11, 2013©2013Elsevier Inc. DOI http://dx.doi.org/10.1016/j.cell.2013.03.031 AUTHOR XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX 1 AFFILIATIONDepartment of XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX Neuroscience and Pharmacology, Rudolf Magnus Institute of Neuroscience, University Medical Center Utrecht, 3584 CG Utrecht, The Netherlands; 2Department of Neuroscience, HHMI, The Johns Hopkins University School of Medicine, Baltimore, MD 21212, USA Axon attraction and repulsion Surround repulsion Selective fasciculation Topographic mapping Self-avoidance Wild-type Dscam1 mutant Sema3A A Retina SC/Tectum EphB EphrinB P D D Mutant T A neuron N P A V V Genomic DNA P EphA EphrinA Slit Exon 4 (12) Exon 6 (48) Exon 9 (33) Exon 17 (2) Netrin Commissural axon guidance Surround repulsion of peripheral Grasshopper CNS axon Retinotectal mapping at the CNS midline nerves in vertebrates fasciculation in vertebrates Drosophila mushroom body XXXXXXXXX NEURITE/CELL Isoneuronal Sema3 Slit Sema1/4-6 Heteroneuronal EphrinA EphrinB FasII Eph Genomic DNA Pcdh-α (14) Pcdh-β (22) Pcdh-γ (22) Variable Con Variable Con Nrp Plexin ** *** Netrin LAMELLIPODIA Con DSCAM ephexin Starburst amacrine cells in mammalian retina Ras-GTP Vav See online version for legend and references. α-chimaerin GEFs/GAPs Robo FARP Ras-GDP LARG RhoGEF Kinases DCC cc0 GTPases PKA cc1 FAK Regulatory Mechanisms See online versionfor??????. Cdc42 GSK3 cc2 Rac PI3K P1 Rho P2 cc3 Abl Proteolytic cleavage P3 Regulation of expression (TF, miRNA, FILOPODIA srGAP Cytoskeleton regulatory proteins multiple isoforms) Sos Trio Pcdh Cis inhibition DOCK180 PAK ROCK Modulation of receptors’ output LIMK Myosin-II Colin Forward and reverse signaling Actin Trafcking and endocytosis NEURONAL GROWTH CONE Microtubules SnapShot: Axon Guidance R. -

Gene Section Short Communication
Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL INIST -CNRS Gene Section Short Communication SSX2IP (synovial sarcoma, X breakpoint 2 interacting protein) Ghazala Khan, Barbara Guinn University of Bedfordshire, Division of Science, Park Square, Luton, Bedfordshire, UK (GK), University of Bedfordshire, Division of Science, Park Square, Luton, Bedfordshire, UK; Cancer Sciences Unit, University of Southampton, Southampton, UK; Department of Haematological Medicine, Kings College, London, UK (BG) Published in Atlas Database: March 2012 Online updated version : http://AtlasGeneticsOncology.org/Genes/SSX2IPID42407ch1p22.html DOI: 10.4267/2042/47489 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence. © 2012 Atlas of Genetics and Cytogenetics in Oncology and Haematology exons however the first one is not translated (de Bruijn Identity et al., 2002). Other names: ADIP Transcription HGNC (Hugo): SSX2IP The gene contains 33 introns. 18 different mRNAs are Location: 1p22.3 produced; 17 spliced and 1 un-spliced form (Thierry- Note Mieg and Thierry-Mieg, 2006). SSX2IP gene encodes the protein SSX2IP which Pseudogene interacts with the cancer-testis antigen SSX2. It is A pseudogene of this gene is found on chromosome 3 thought that SSX2IP regulates the function of SSX2 in (provided by RefSeq, Oct 2009 from Entrez Gene). the testes and malignant cells. The rodent equivalent is known as afadin DIL domain-interacting protein (ADIP) and the chicken orthologue is called clock- Protein controlled gene (LCG) (Breslin et al., 2007). Note SSX2IP was discovered due to its interaction with DNA/RNA SSX2 in a yeast two-hybrid system and believed to regulate the function of SSX2 in the testes and Note malignant cells (de Bruijn et al., 2002). -

Regulation of the Mammalian Target of Rapamycin Complex 2 (Mtorc2)
Regulation of the Mammalian Target Of Rapamycin Complex 2 (mTORC2) Inauguraldissertation Zur Erlangung der Würde eines Doktors der Philosophie vorgelegt der Philosophisch-Naturwissenschaftlichen Fakultät der Universität Basel von Klaus-Dieter Molle aus Heilbronn, Deutschland Basel, 2006 Genehmigt von der Philosophisch-Naturwissenschaftlichen Fakultät Auf Antrag von Prof. Michael N. Hall und Prof. Markus Affolter. Basel, den 21.11.2006 Prof. Hans-Peter Hauri Dekan Summary The growth controlling mammalian Target of Rapamycin (mTOR) is a conserved Ser/Thr kinase found in two structurally and functionally distinct complexes, mTORC1 and mTORC2. The tumor suppressor TSC1-TSC2 complex inhibits mTORC1 by acting on the small GTPase Rheb, but the role of TSC1-TSC2 and Rheb in the regulation of mTORC2 is unclear. Here we examined the role of TSC1-TSC2 in the regulation of mTORC2 in human embryonic kidney 293 cells. Induced knockdown of TSC1 and TSC2 (TSC1/2) stimulated mTORC2-dependent actin cytoskeleton organization and Paxillin phosphorylation. Furthermore, TSC1/2 siRNA increased mTORC2-dependent Ser473 phosphorylation of plasma membrane bound, myristoylated Akt/PKB. This suggests that loss of Akt/PKB Ser473 phosphorylation in TSC mutant cells, as reported previously, is due to inhibition of Akt/PKB localization rather than inhibition of mTORC2 activity. Amino acids and overexpression of Rheb failed to stimulate mTORC2 signaling. Thus, TSC1-TSC2 also inhibits mTORC2, but possibly independently of Rheb. Our results suggest that mTORC2 hyperactivation may contribute to the pathophysiology of diseases such as cancer and Tuberous Sclerosis Complex. i Acknowledgement During my PhD studies in the Biozentrum I received a lot of support from many people around me who I mention here to express my gratefulness. -

The Atypical Guanine-Nucleotide Exchange Factor, Dock7, Negatively Regulates Schwann Cell Differentiation and Myelination
The Journal of Neuroscience, August 31, 2011 • 31(35):12579–12592 • 12579 Cellular/Molecular The Atypical Guanine-Nucleotide Exchange Factor, Dock7, Negatively Regulates Schwann Cell Differentiation and Myelination Junji Yamauchi,1,3,5 Yuki Miyamoto,1 Hajime Hamasaki,1,3 Atsushi Sanbe,1 Shinji Kusakawa,1 Akane Nakamura,2 Hideki Tsumura,2 Masahiro Maeda,4 Noriko Nemoto,6 Katsumasa Kawahara,5 Tomohiro Torii,1 and Akito Tanoue1 1Department of Pharmacology and 2Laboratory Animal Resource Facility, National Research Institute for Child Health and Development, Setagaya, Tokyo 157-8535, Japan, 3Department of Biological Sciences, Tokyo Institute of Technology, Midori, Yokohama 226-8501, Japan, 4IBL, Ltd., Fujioka, Gumma 375-0005, Japan, and 5Department of Physiology and 6Bioimaging Research Center, Kitasato University School of Medicine, Sagamihara, Kanagawa 252-0374, Japan In development of the peripheral nervous system, Schwann cells proliferate, migrate, and ultimately differentiate to form myelin sheath. In all of the myelination stages, Schwann cells continuously undergo morphological changes; however, little is known about their underlying molecular mechanisms. We previously cloned the dock7 gene encoding the atypical Rho family guanine-nucleotide exchange factor (GEF) and reported the positive role of Dock7, the target Rho GTPases Rac/Cdc42, and the downstream c-Jun N-terminal kinase in Schwann cell migration (Yamauchi et al., 2008). We investigated the role of Dock7 in Schwann cell differentiation and myelination. Knockdown of Dock7 by the specific small interfering (si)RNA in primary Schwann cells promotes dibutyryl cAMP-induced morpholog- ical differentiation, indicating the negative role of Dock7 in Schwann cell differentiation. It also results in a shorter duration of activation of Rac/Cdc42 and JNK, which is the negative regulator of myelination, and the earlier activation of Rho and Rho-kinase, which is the positive regulator of myelination. -

Epha2 Proteomics in Human Keratinocytes Reveals a Novel Association with Afadin and Epidermal Tight Junctions Bethany E
© 2017. Published by The Company of Biologists Ltd | Journal of Cell Science (2017) 130, 111-118 doi:10.1242/jcs.188169 SPECIAL ISSUE: 3D CELL BIOLOGY SHORT REPORT EphA2 proteomics in human keratinocytes reveals a novel association with afadin and epidermal tight junctions Bethany E. Perez White1, Rosa Ventrella1, Nihal Kaplan1, Calvin J. Cable1, Paul M. Thomas2,3 and Spiro Getsios1,4,*,‡ ABSTRACT EphA2 increased susceptibility to chemically-induced skin EphA2 is a receptor tyrosine kinase that helps to maintain epidermal carcinogenesis (Guo et al., 2006), whereas ephrin-targeting of tissue homeostasis. A proximity-dependent biotin identification (BioID) EphA2 enhanced keratinocyte adhesion and differentiation (Lin approach was used to identify proteins in close proximity to EphA2 et al., 2010; Walsh and Blumenberg, 2011). EphA2 can positively within primary human keratinocytes and three-dimensional (3D) or negatively regulate intercellular junctions, including tight reconstituted human epidermis (RHE) cultures to map a putative junctions (Zhou et al., 2011; Tanaka et al., 2005; Larson et al., protein interaction network for this membrane receptor that exhibits a 2008; Miao et al., 2014; Miura et al., 2009) that contribute to skin polarized distribution in stratified epithelia. Although a subset of known barrier function (Niessen, 2007). Importantly, EphA2 is expressed EphA2 interactors were identified in the BioID screen, >97% were in a differentiation-dependent, polarized manner within human uniquely detected in keratinocytes with over 50% of these vicinal epidermis (Fig. 1A). proteins only present in 3D human epidermal culture. Afadin (AFDN), a We adapted the method of unbiased, proximity-dependent biotin cytoskeletal and junction-associated protein, was present in 2D and 3D identification (BioID) (Roux et al., 2012) to identify near neighbors keratinocyte cultures, and validated as a so-far-unknown EphA2- of EphA2 in normal human epidermal keratinocytes (NHEKs) 2+ interacting protein. -

Afadin, a Ras/Rap Effector That Controls Cadherin Function, Promotes Spine and Excitatory Synapse Density in the Hippocampus
The Journal of Neuroscience, January 4, 2012 • 32(1):99–110 • 99 Development/Plasticity/Repair Afadin, A Ras/Rap Effector That Controls Cadherin Function, Promotes Spine and Excitatory Synapse Density in the Hippocampus Gerard M. J. Beaudoin III,1 Claude M. Schofield,2* Tulip Nuwal,3* Keling Zang,1 Erik M. Ullian,1,2† Bo Huang,3† and Louis F. Reichardt1,4 Departments of 1Physiology, 2Opthamology, 3Pharmaceutical Chemistry, and 4Biochemistry and Biophysics, University of California, San Francisco, San Francisco, California 94158 Many molecules regulate synaptogenesis, but intracellular signaling pathways required for their functions are poorly understood. Afadin is a Rap-regulated, actin-binding protein that promotes cadherin complex assembly as well as binding many other cell adhesion mole- cules and receptors. To examine its role in mediating synaptogenesis, we deleted afadin (mllt1), using a conditional allele, in postmitotic hippocampal neurons. Consistent with its role in promoting cadherin recruitment, afadin deletion resulted in 70% fewer and less intense N-cadherin puncta with similar reductions of -catenin and ␣N-catenin puncta densities and 35% reduction in EphB2 puncta density. Its absence also resulted in 40% decreases in spine and excitatory synapse densities in the stratum radiatum of CA1, as determined by morphology, apposition of presynaptic and postsynaptic markers, and synaptic transmission. The remaining synapses appeared to function normally. Thus, afadin is a key intracellular signaling molecule for cadherin recruitment and is necessary for spine and synapse formation in vivo. Introduction 2007). In Drosophila, absence of N-cadherin prevents normal Synapses are formed at contacts between neurons as a result of synapse formation between photoreceptor axons and their tar- intercellular interactions that result in stepwise assembly of syn- gets (Clandinin and Feldheim, 2009). -

Rap1 Acts Via Multiple Mechanisms to Position Canoe/Afadin and Adherens Junctions and Mediate Apical-Basal Polarity Establishment
bioRxiv preprint doi: https://doi.org/10.1101/170977; this version posted July 31, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Rap1 acts via multiple mechanisms to position Canoe/Afadin and adherens junctions and mediate apical-basal polarity establishment Teresa T. Bonello1, Kia Z. Perez-Vale2, Kaelyn D. Sumigray3, and Mark Peifer1,2,3* 1 Department of Biology, University of North Carolina at Chapel Hill, CB#3280, Chapel Hill, NC 27599-3280, USA 2 Curriculum in Genetics and Molecular Biology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA 3 Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA Running Title Active Rap1 positions Canoe and AJs 6950 words * To whom correspondence should be addressed Email: [email protected] Phone: (919) 962-2272 Abbreviations used: α-cat, alpha-catenin; β-cat, beta-catenin; AJ, adherens junction; Arm, Armadillo; Baz, BazooKa; CA, constitutively active; Cno, Canoe; DE-cad, Drosophila E-cadherin; Dzy, Dizzy; GAP, GTPase activating protein; GDP, guanosine diphosphate; GEF, guanine nucleotide exchange factor; GFP, green fluorescent protein; GTP, guanosine triphosphate; IF, immunofluorescence; MIP, maximum intensity projection; RA, Ras-associated; RFP, red fluorescent protein; SAJ, spot adherens junction; shRNA, short hairpin RNA; TCJ, tricellular junction; WT, wildtype 1 bioRxiv preprint doi: https://doi.org/10.1101/170977; this version posted July 31, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. -

Identification of Two Signaling Submodules Within the Crkii/ELMO
Cell Death and Differentiation (2007) 14, 963–972 & 2007 Nature Publishing Group All rights reserved 1350-9047/07 $30.00 www.nature.com/cdd Identification of two signaling submodules within the CrkII/ELMO/Dock180 pathway regulating engulfment of apoptotic cells A-C Tosello-Trampont1,4, JM Kinchen1,2,4, E Brugnera1,3, LB Haney1, MO Hengartner2 and KS Ravichandran*,1 Removal of apoptotic cells is a dynamic process coordinated by ligands on apoptotic cells, and receptors and other signaling proteins on the phagocyte. One of the fundamental challenges is to understand how different phagocyte proteins form specific and functional complexes to orchestrate the recognition/removal of apoptotic cells. One evolutionarily conserved pathway involves the proteins cell death abnormal (CED)-2/chicken tumor virus no. 10 (CT10) regulator of kinase (Crk)II, CED-5/180 kDa protein downstream of chicken tumor virus no. 10 (Crk) (Dock180), CED-12/engulfment and migration (ELMO) and MIG-2/RhoG, leading to activation of the small GTPase CED-10/Rac and cytoskeletal remodeling to promote corpse uptake. Although the role of ELMO : Dock180 in regulating Rac activation has been well defined, the function of CED-2/CrkII in this complex is less well understood. Here, using functional studies in cell lines, we observe that a direct interaction between CrkII and Dock180 is not required for efficient removal of apoptotic cells. Similarly, mutants of CED-5 lacking the CED-2 interaction motifs could rescue engulfment and migration defects in CED-5 deficient worms. Mutants of CrkII and Dock180 that could not biochemically interact could colocalize in membrane ruffles. -

Vinexin Family (SORBS) Proteins Play Different Roles in Stiffness- Sensing and Contractile Force Generation Takafumi Ichikawa1,2, Masahiro Kita1, Tsubasa S
© 2017. Published by The Company of Biologists Ltd | Journal of Cell Science (2017) 130, 3517-3531 doi:10.1242/jcs.200691 RESEARCH ARTICLE Vinexin family (SORBS) proteins play different roles in stiffness- sensing and contractile force generation Takafumi Ichikawa1,2, Masahiro Kita1, Tsubasa S. Matsui3,4, Ayaka Ichikawa Nagasato1, Tomohiko Araki3, Shian-Huey Chiang5, Takuhito Sezaki1, Yasuhisa Kimura1, Kazumitsu Ueda1,2, Shinji Deguchi3,4, Alan R. Saltiel5,* and Noriyuki Kioka1,2,‡ ABSTRACT generating actin stress fibers (SFs) (Geiger et al., 2001). This Vinexin, c-Cbl associated protein (CAP) and Arg-binding protein 2 ‘ ’ (ArgBP2) constitute an adaptor protein family called the vinexin mechanical linkage acts as a molecular clutch to transmit the force (SORBS) family that is targeted to focal adhesions (FAs). Although derived from non-muscle myosin-II-dependent contraction to the numerous studies have focused on each of the SORBS proteins and ECM. Cells on more rigid substrates exert greater contractile forces partially elucidated their involvement in mechanotransduction, a than those on soft substrates (Hoffman et al., 2011; Roca-Cusachs comparative analysis of their function has not been well addressed. et al., 2012; LaCroix et al., 2015). These alterations can lead to Here, we established mouse embryonic fibroblasts that individually stiffness-dependent biochemical signals. Among the numerous FA scaffolding proteins, vinculin is one of expressed SORBS proteins and analysed their functions in an ‘ ’ identical cell context. Both vinexin-α and CAP co-localized with the main clutch molecules that can regulate force transmission. vinculin at FAs and promoted the appearance of vinculin-rich FAs, Vinculin consists of an N-terminal head region and a C-terminal tail α region separated by a flexible proline-rich linker region (Bakolitsa whereas ArgBP2 co-localized with -actinin at the proximal end of – FAs and punctate structures on actin stress fibers (SFs), and induced et al., 2004; Borgon et al., 2004). -

Disassembly of the Dying: Mechanisms and Functions
Review Disassembly of the Dying: Mechanisms and Functions 1 1, Georgia K. Atkin-Smith and Ivan K.H. Poon * The disassembly of an apoptotic cell into subcellular fragments, termed Trends apoptotic bodies (ApoBDs), is a hallmark of apoptosis. Although the genera- Apoptotic cell disassembly is a highly tion of ApoBDs is generally understood as being stochastic, it is becoming complex process regulated by a series of well-coordinated morphological increasingly clear that ApoBD formation is a highly regulated process involv- steps including apoptotic membrane ing distinct morphological steps and molecular factors. Functionally, ApoBDs blebbing, apoptotic protrusion forma- fi could facilitate the ef cient clearance of apoptotic material by surrounding tion, and fragmentation. phagocytes as well as mediate the transfer of biomolecules including micro- Plasma-membrane blebbing is not the RNAs and proteins between cells to aid in intercellular communications. sole process required for apoptotic Therefore, the formation of ApoBDs is an important process downstream body (ApoBD) formation, but mem- brane protrusions including microtu- from apoptotic cell death. We discuss here the mechanisms and functions bule spikes, apoptopodia, and of apoptotic cell disassembly. beaded apoptopodia may act in con- cert to aid the generation of ApoBDs. Cell Disassembly as a Key Downstream Process of Apoptotic Cell Death The mechanism of how apoptotic cells Billions of cells undergo apoptosis (a form of programmed cell death) daily as part of disassembly can determine the quantity and quality (size and contents) of physiological homeostasis [1]. At later stages of apoptosis, some cell types can generate ApoBDs. subcellular (1–5 mm) membrane-bound extracellular vesicles termed apoptotic bodies – (ApoBDs, see Glossary) [1 3]. -

Kobe University Repository : Thesis
Kobe University Repository : Thesis Afadin Regulates Puncta Adherentia Junction Formation and 学位論文題目 Presynaptic Differentiation in Hippocampal Neurons(アファディンは海 Title 馬ニューロンにおいてアドへレンスジャンクションの形成と前シナプ スの分化を調節している) 氏名 Toyoshima, Daisaku Author 専攻分野 博士(医学) Degree 学位授与の日付 2014-03-25 Date of Degree 公開日 2015-03-01 Date of Publication 資源タイプ Thesis or Dissertation / 学位論文 Resource Type 報告番号 甲第6188号 Report Number 権利 Rights JaLCDOI URL http://www.lib.kobe-u.ac.jp/handle_kernel/D1006188 ※当コンテンツは神戸大学の学術成果です。無断複製・不正使用等を禁じます。著作権法で認められている範囲内で、適切にご利用ください。 PDF issue: 2021-09-26 Afadin Regulates Puncta Adherentia Junction Formation and Presynaptic Differentiation in Hippocampal Neurons アファディンは海馬ニューロンにおいてアドへレンスジャンクション の形成と前シナプスの分化を調節している 豊嶋大作, 萬代研二, 丸尾知彦、Irwan Supriyanto、 富樫英、井上貴仁、森正弘、高井義美 神戸大学大学院医学研究科医科学専攻 小児科学 (指導教員:飯島一誠 教授) 豊嶋大作 Key words: Afadin, puncta adherentia junction, presynapse, hippocampal neuron Manuscript Click here to download Manuscript: Toyoshima_text_revised_2nd_final.pdf 1 Afadin Regulates Puncta Adherentia Junction Formation and 2 Presynaptic Differentiation in Hippocampal Neurons 3 4 Daisaku Toyoshima1,3,4, Kenji Mandai1,3, Tomohiko Maruo1,3, Irwan Supriyanto2,3, 5 Hideru Togashi1,3, Takahito Inoue1,3, Masahiro Mori2,3 & Yoshimi Takai1,3 6 7 1. Department of Biochemistry and Molecular Biology, Kobe University Graduate 8 School of Medicine, Kobe, Hyogo 650-0047, Japan. 9 2. Faculty of Health Sciences, Kobe University Graduate School of Health Sciences, 10 Kobe, Hyogo 654-0142, Japan. 11 3. CREST, Japan Science and Technology Agency, Kobe, Hyogo 650-0047, Japan. 12 4. Present address: Department of Pediatrics, Kobe University Graduate School of 13 Medicine, Kobe, Hyogo 650-0017, Japan. 14 15 Correspondence should be addressed to Y.T. ([email protected]), K.M. 16 ([email protected]). -

Snapshot: Axon Guidance II Alex L
SnapShot: Axon Guidance II Alex L. Kolodkin1 and R. Jeroen Pasterkamp2 1Department of Neuroscience, HHMI, The Johns Hopkins University School of Medicine, Baltimore, MD 21212, USA 2Department of Neuroscience and Pharmacology, Rudolf Magnus Institute of Neuroscience, University Medical Center Utrecht, 3584 CG Utrecht, The Netherlands Axon guidance protein Chemotrophic effect Ligand-binding receptor Receptor processing Coreceptor Receptor signaling COFILIN, LIMK1, PI3K, BMP SE Repulsion BMP-RIB, BMP-RII - - SMAD1, SMAD6 DRAXIN SE Repulsion DCC - - - a2-CHIMAERIN, EPHEXIN-1, EPHRINA GPI Repulsion EPHA ADAM10 - NCK1/2, RAC1, RHOA, SPAR, VAV2/3 NCK1, PAK, p120GAP, RHOA, EPHRINB TM Repulsion EPHB MMP2/9 - ROCK, VAV2/3 TM Repulsion EPHRINA ADAM10 P75 FYN EPHA Attraction EPHRINA - RET - CDC42, DOCK180, GRB4, NCK2, EPHB TM Repulsion EPHRINB ADAM10, PS1 - PAK, RAC1 FGF SE Attraction FGFR1 - - - CDC42, DOCK180, ENA/VASP, APP, HSPG, ERK1/2, FAK, FYN, NCK1, N- SE Attraction DCC ADAM10, PS1 ROBO1 WASP, PAK, PI3K, PIP2, PKC, NETRIN RAC1, RHOA, TRIO, TRP Attraction NEOGENIN - - - Repulsion UNC5 - (DCC) FAK, SHP2, SRC FAK, LARG, LMO4, MYOIIA, RGM GPI Repulsion NEOGENIN TACE, PS1 UNC5 p120GAP, PKC, RAS, RHOA PLEXINA TM Repulsion SEMA1a - - PEBBLE, RHO, p190RHOGAP 14-3-3e, GYC76C, MICAL, SEMA1 TM Repulsion PLEXINA - OTK NERVY, PKA SE Repulsion PLEXINB - - RAC, RHO SEMA2 Attraction PLEXINB - - - AKT, COFILIN, CDK5, CRMP, FARP, NRP1/2, CAMs, SE Repulsion PLEXINA1-4 CALPAIN1 FYN, GSK3b, LIMK1, PI3K, RAC1, RTKs, ROBO SEMA3 RAP1, RAS, RND Attraction - - NRP1/2,