Anaerobic Microbial LCFA Degradation in Bioreactors D
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Fatty Acid Diets: Regulation of Gut Microbiota Composition and Obesity and Its Related Metabolic Dysbiosis
International Journal of Molecular Sciences Review Fatty Acid Diets: Regulation of Gut Microbiota Composition and Obesity and Its Related Metabolic Dysbiosis David Johane Machate 1, Priscila Silva Figueiredo 2 , Gabriela Marcelino 2 , Rita de Cássia Avellaneda Guimarães 2,*, Priscila Aiko Hiane 2 , Danielle Bogo 2, Verônica Assalin Zorgetto Pinheiro 2, Lincoln Carlos Silva de Oliveira 3 and Arnildo Pott 1 1 Graduate Program in Biotechnology and Biodiversity in the Central-West Region of Brazil, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; [email protected] (D.J.M.); [email protected] (A.P.) 2 Graduate Program in Health and Development in the Central-West Region of Brazil, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; pri.fi[email protected] (P.S.F.); [email protected] (G.M.); [email protected] (P.A.H.); [email protected] (D.B.); [email protected] (V.A.Z.P.) 3 Chemistry Institute, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; [email protected] * Correspondence: [email protected]; Tel.: +55-67-3345-7416 Received: 9 March 2020; Accepted: 27 March 2020; Published: 8 June 2020 Abstract: Long-term high-fat dietary intake plays a crucial role in the composition of gut microbiota in animal models and human subjects, which affect directly short-chain fatty acid (SCFA) production and host health. This review aims to highlight the interplay of fatty acid (FA) intake and gut microbiota composition and its interaction with hosts in health promotion and obesity prevention and its related metabolic dysbiosis. -

Anaerobic Microbial LCFA Degradation in Bioreactors
Presented in Session PP3A – Bio-electrochemical Processes 11th IWA World Congress on Anaerobic Digestion 23-27 September 2007 Brisbane, Australia Anaerobic microbial LCFA degradation in bioreactors D.Z. Sousa*, M.A. Pereira*, J.I. Alves*, H. Smidt**, A.J.M. Stams**, M.M. Alves* * Institute for Biotechnology and Bioengineering, Center for Biological Engineering, University of Minho, Campus de Gualtar, 4710-057 Braga, Portugal (E-mail: [email protected]; [email protected]) ** Laboratory of Microbiology, Wageningen University, Hesselink van Suchtelenweg 4, 6703 CT Wageningen, The Netherlands Abstract This paper reviews recent results obtained on long-chain fatty acids (LCFA) anaerobic degradation. Two LCFA were used as model substrates: oleate, a mono-unsaturated LCFA, and palmitate, a saturated LCFA, both abundant in LCFA-rich wastewaters. 16S rRNA gene analysis of sludge samples submitted to continuous oleate- and palmitate-feeding followed by batch degradation of the accumulated LCFA demonstrated that bacterial communities were dominated by members of the Clostridiaceae and Syntrophomonadaceae families. Archaeal populations were mainly comprised of hydrogen-consuming microorganisms belonging to the genus Methanobacterium, and acetate- utilizers from the genera Methanosaeta and Methanosarcina. Enrichment cultures growing on oleate and palmitate, in the absence or presence of sulfate, gave more insight into the major players involved in the degradation of unsaturated and saturated LCFA. Syntrophomonas-related species were identified as predominant microorganisms in all the enrichment cultures. Microorganisms clustering within the family Syntrophobacteraceae were identified in the methanogenic and sulfate-reducing enrichments growing on palmitate. Distinct bacterial consortia were developed in oleate and palmitate enrichments, and observed differences might be related to the different degrees of saturation of these two LCFA. -

From Sporulation to Intracellular Offspring Production: Evolution
FROM SPORULATION TO INTRACELLULAR OFFSPRING PRODUCTION: EVOLUTION OF THE DEVELOPMENTAL PROGRAM OF EPULOPISCIUM A Dissertation Presented to the Faculty of the Graduate School of Cornell University In Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy by David Alan Miller January 2012 © 2012 David Alan Miller FROM SPORULATION TO INTRACELLULAR OFFSPRING PRODUCTION: EVOLUTION OF THE DEVELOPMENTAL PROGRAM OF EPULOPISCIUM David Alan Miller, Ph. D. Cornell University 2012 Epulopiscium sp. type B is an unusually large intestinal symbiont of the surgeonfish Naso tonganus. Unlike most other bacteria, Epulopiscium sp. type B has never been observed to undergo binary fission. Instead, to reproduce, it forms multiple intracellular offspring. We believe this process is related to endospore formation, an ancient and complex developmental process performed by certain members of the Firmicutes. Endospore formation has been studied for over 50 years and is best characterized in Bacillus subtilis. To study the evolution of endospore formation in the Firmicutes and the relatedness of this process to intracellular offspring formation in Epulopiscium, we have searched for sporulation genes from the B. subtilis model in all of the completed genomes of members of the Firmicutes, in addition to Epulopiscium sp. type B and its closest relative, the spore-forming Cellulosilyticum lentocellum. By determining the presence or absence of spore genes, we see the evolution of endospore formation in closely related bacteria within the Firmicutes and begin to predict if 19 previously characterized non-spore-formers have the genetic capacity to form a spore. We can also map out sporulation-specific mechanisms likely being used by Epulopiscium for offspring formation. -
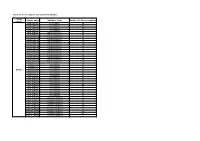
Table S1: List of Samples Included in the Analysis
Table S1: list of samples included in the analysis Study Sample name Inhibitory status Number of days at sampling number DNA.0P2T4 No inhibition 29 DNA.0P2T6 No inhibition 57 DNA.10P2T4 No inhibition 29 DNA.10P2T6 No inhibition 57 DNA.75P2T4 Phenol inhibition 29 DNA.75P2T6 Phenol inhibition 57 DNA.100P2T4 Phenol inhibition 29 DNA.100P2T6 Phenol inhibition 57 DNA.125P1T4 Phenol inhibition 29 DNA.125P1T6 Phenol inhibition 57 DNA.125P2T4 Phenol inhibition 29 DNA.125P2T6 Phenol inhibition 57 DNA.125P3T4 Phenol inhibition 29 DNA.125P3T6 Phenol inhibition 57 DNA.150P2T4 Phenol inhibition 29 DNA.150P2T6 Phenol inhibition 57 DNA.200P2T4 Phenol inhibition 29 DNA.200P2T6 Phenol inhibition 57 DNA.0N2T4 No inhibition 29 DNA.0N2T5 No inhibition 42 DNA.0N2T6 No inhibition 57 Study 1 DNA.5N2T4 No inhibition 29 DNA.5N2T5 No inhibition 42 DNA.5N2T6 No inhibition 57 DNA.10N2T4 No inhibition 29 DNA.10N2T5 No inhibition 42 DNA.10N2T6 No inhibition 57 DNA.15N2T4 No inhibition 29 DNA.15N2T5 No inhibition 42 DNA.15N2T6 No inhibition 57 DNA.25N2T4 No inhibition 29 DNA.25N2T5 No inhibition 42 DNA.25N2T6 No inhibition 57 DNA.75N2T4 Ammonia inhibition 29 DNA.75N2T5 Ammonia inhibition 42 DNA.75N2T6 Ammonia inhibition 57 DNA.100N2T4 Ammonia inhibition 29 DNA.100N2T5 Ammonia inhibition 42 DNA.100N2T6 Ammonia inhibition 57 DNA.250N2T4 Ammonia inhibition 29 DNA.250N2T5 Ammonia inhibition 42 DNA.250N2T6 Ammonia inhibition 57 nono2T3 No inhibition 16 noN2T4 Ammonia inhibition 23 noN2T8 Ammonia inhibition 60 noN2T9 Ammonia inhibition 85 noPhi2T4 Phenol inhibition 23 noPhi2T5 -

Syntrophic Butyrate and Propionate Oxidation Processes 491
Environmental Microbiology Reports (2010) 2(4), 489–499 doi:10.1111/j.1758-2229.2010.00147.x Minireview Syntrophic butyrate and propionate oxidation processes: from genomes to reaction mechanismsemi4_147 489..499 Nicolai Müller,1† Petra Worm,2† Bernhard Schink,1* a cytoplasmic fumarate reductase to drive energy- Alfons J. M. Stams2 and Caroline M. Plugge2 dependent succinate oxidation. Furthermore, we 1Faculty for Biology, University of Konstanz, D-78457 propose that homologues of the Thermotoga mar- Konstanz, Germany. itima bifurcating [FeFe]-hydrogenase are involved 2Laboratory of Microbiology, Wageningen University, in NADH oxidation by S. wolfei and S. fumaroxidans Dreijenplein 10, 6703 HB Wageningen, the Netherlands. to form hydrogen. Summary Introduction In anoxic environments such as swamps, rice fields In anoxic environments such as swamps, rice paddy fields and sludge digestors, syntrophic microbial communi- and intestines of higher animals, methanogenic commu- ties are important for decomposition of organic nities are important for decomposition of organic matter to matter to CO2 and CH4. The most difficult step is the CO2 and CH4 (Schink and Stams, 2006; Mcinerney et al., fermentative degradation of short-chain fatty acids 2008; Stams and Plugge, 2009). Moreover, they are the such as propionate and butyrate. Conversion of these key biocatalysts in anaerobic bioreactors that are used metabolites to acetate, CO2, formate and hydrogen is worldwide to treat industrial wastewaters and solid endergonic under standard conditions and occurs wastes. Different types of anaerobes have specified only if methanogens keep the concentrations of these metabolic functions in the degradation pathway and intermediate products low. Butyrate and propionate depend on metabolite transfer which is called syntrophy degradation pathways include oxidation steps of (Schink and Stams, 2006). -

Biotechnology for Biofuels
Biotechnology for Biofuels This Provisional PDF corresponds to the article as it appeared upon acceptance. Fully formatted PDF and full text (HTML) versions will be made available soon. Comparative metagenomics of biogas-producing microbial communities from production-scale biogas plants operating under wet or dry fermentation conditions Biotechnology for Biofuels (2015)8:14Sample doi:10.1186/s13068-014-0193-8 Yvonne Stolze ([email protected]) Martha Zakrzewski ([email protected]) Irena Maus ([email protected]) Felix Eikmeyer ([email protected]) Sebastian Jaenicke ([email protected]) Nils Rottmann ([email protected]) Clemens Siebner ([email protected]) Alfred Pühler ([email protected]) Andreas Schlüter ([email protected]) Sample ISSN 1754-6834 Article type Research article Submission date 29 September 2014 Acceptance date 22 December 2014 Article URL http://dx.doi.org/10.1186/s13068-014-0193-8 Like all articles in BMC journals, this peer-reviewed article can be downloaded, printed and distributed freely for any purposes (see copyright notice below). Articles in BMC journals are listed in PubMed and archived at PubMed Central. For information about publishing your research in BMC journals or any BioMed Central journal, go to http://www.biomedcentral.com/info/authors/ © 2015 Stolze et al.; licensee BioMed Central. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly credited. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated. -

Analysis of Microbial Community Structure of Pit Mud for Chinese Strong-Flavor Liquor Fermentation Using Next Generation DNA Sequencing of Full-Length 16S Rrna
bioRxiv preprint doi: https://doi.org/10.1101/380949; this version posted July 31, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Analysis of microbial community structure of pit mud for Chinese strong-flavor liquor fermentation using next generation DNA sequencing of full-length 16S rRNA Zuolin Liu1†, Ying Han1†, Junwei Li1,2, Runjie Cao2, Hongkui He2*, Anjun Li2 and Zhizhou Zhang1* 1School of Chemical Engineering and Chemistry, Harbin Institute of Technology, Harbin, China 150006 2The Center for Solid-state Fermentation Engineering of Anhui Province, The Anhui GujingTribute Liquor Ltd, Bozhou, Anhui, China, 236800 † *The corresponding author: [email protected]; [email protected] Equal contribution Contact: Zhizhou Zhang, PhD Prof [email protected] 86-631-5683176 Abstract. The pit is the necessary bioreactor for brewing process of Chinese strong-flavor liquor. Pit mud in pits contains a large number of microorganisms and is a complex ecosystem. The analysis of bacterial flora in pit mud is of great significance to understand liquor fermentation mechanisms. To overcome taxonomic limitations of short reads in 16S rRNA variable region sequencing, we used high-throughput DNA sequencing of near full-length 16S rRNA gene to analyze microbial compositions of different types of pit mud that produce different qualities of strong-flavor liquor. The results showed that the main species in pit mud were Pseudomonas extremaustralis 14-3, Pseudomonas veronii, Serratia marcescens WW4, and Clostridium leptum in Ruminiclostridium. -

The Core Populations and Co-Occurrence Patterns Of
Rui et al. Biotechnol Biofuels (2015) 8:158 DOI 10.1186/s13068-015-0339-3 RESEARCH Open Access The core populations and co‑occurrence patterns of prokaryotic communities in household biogas digesters Junpeng Rui1,2†, Jiabao Li1,2†, Shiheng Zhang1,2, Xuefeng Yan1,2, Yuanpeng Wang3 and Xiangzhen Li1,2* Abstract Background: Household biogas digesters are widely used to harvest energy in rural areas of developing countries. Understanding core prokaryotic communities, their co-occurrence patterns, and their relationships to environmental factors is important to manage these small-scale anaerobic digestion systems effectively. In this study, 43 household biogas digesters were collected across eight provinces in China. Prokaryotic communities were investigated using 454 pyrosequencing of 16S rRNA genes. Results: Fourteen core genera and ten core OTUs were identified in household biogas digesters. They were mainly affiliated with the phylum Firmicutes, Synergistetes, Actinobacteria, Chloroflexi, and Spirochaetes. Core prokaryotic genera were mainly composed of Clostridium, Clostridium XI, Syntrophomonas, Cloacibacillus, Sedimentibacter, and Turicibacter. Prokaryotic communities in the 43 samples were clearly divided into two clusters. Cluster I was domi- nated by Clostridium, while Cluster II was dominated by members of Spirochaetes, Bacteroidales, Clostridia, and abundant syntrophs and methanogens. NH4+-N and COD contributed significantly to the assembly of the prokaryotic community in Cluster I, while NH4+-N, pH, and phosphate contributed significantly to Cluster II. Correlation-based network analysis showed that the prokaryotic communities in the biogas digesters were dominated by some func- tional modules. Cluster I was dominated by acetotrophic methanogenic modules and the Clostridium-driven primary fermentation module, while the network of Cluster II was dominated by hydrogenotrophic and acetogenic methano- genesis modules and multi-group-driven (Spirochaetes, Bacteroidales, and Clostridia) primary fermentation modules. -

Elucidating Syntrophic Butyrate-Degrading Populations in Anaerobic Digesters
bioRxiv preprint doi: https://doi.org/10.1101/563387; this version posted February 28, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Elucidating syntrophic butyrate-degrading populations in anaerobic digesters 2 using stable isotope-informed genome-resolved metagenomics 3 4 Ryan M. Ziels1,2,#, Masaru K. Nobu3, Diana Z. Sousa4 5 6 7 1 Department of Civil Engineering, University of British Columbia, Vancouver, Canada 8 2 Department of Civil and Environmental Engineering, University Washington, Seattle, USA 9 3 Bioproduction Research Institute, National Institute of Advanced Industrial Science and 10 Technology, Tsukuba, Japan 11 4 Laboratory of Microbiology, Wageningen University & Research, Wageningen, Netherlands 12 13 #Corresponding Author: 14 Email: [email protected] 15 16 Running Title: DNA-SIP Metagenomics of Anaerobic Butyrate-Degraders 17 1 bioRxiv preprint doi: https://doi.org/10.1101/563387; this version posted February 28, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 18 Abstract: 19 Linking the genomic content of uncultivated microbes to their metabolic functions remains a 20 critical challenge in microbial ecology. Resolving this challenge has implications for improving 21 our management of key microbial interactions in biotechnologies such as anaerobic digestion, 22 which relies on slow-growing syntrophic and methanogenic communities to produce renewable 23 methane from organic waste. -

Nutritional Features of Syntrophomonas Wolfei P
APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Oct. 1990, p. 3223-3224 Vol. 56, No. 10 0099-2240/90/103223-02$02.00/0 Copyright © 1990, American Society for Microbiology Nutritional Features of Syntrophomonas wolfei P. SHAWN BEATY AND MICHAEL J. McINERNEY* Department ofBotany and Microbiology, University of Oklahoma, Norman, Oklahoma 73019-0245 Received 27 March 1990/Accepted 18 July 1990 Syntrophomonas wolfei subsp. wolfei grew poorly in a defined medium with crotonate as the energy source in the absence of rumen fluid. Thiamine, lipoic acid, biotin, cyanocobalamin, and para-aminobenzoic acid were required for growth comparable to that obtained with the rumen fluid-based medium. Iron and cobalt were also required for the growth of S. wolfei in the chemically defined medium. Propionate and longer-chain fatty acids are important in medium lacking a particular compound before it was intermediates in the conversion of organic matter to CH4 and concluded that the compound was not required for the CO2 (4). These compounds are degraded to methanogenic growth. Growth was monitored spectrophotometrically at substrates (acetate, H2, and formate) by syntrophic bacteria 600 nm, and growth rates were calculated from changes in which require the presence of methanogens or other H2- absorbance (6). using bacteria to maintain low concentrations of H2 for the The pure culture of S. wolfei grew at a specific growth rate degradation offatty acids to be thermodynamically favorable of 0.039/h in crotonate basal medium which contained rumen (4). Syntrophomonas wolfei subsp. wolfei is an anaerobic fluid. Decreasing the rumen fluid concentration of this me- bacterium that a-oxidizes short-chain saturated fatty acids, dium from 5 to 0.5% (vol/vol) did not affect the rate of four to eight carbons in length, to acetate, propionate (from growth or the final absorbance (0.45 to 0.5) of the culture odd-numbered fatty acids), and H2 only when grown in (data not shown). -

Research Article Archaea and Bacteria Acclimate to High Total Ammonia in a Methanogenic Reactor Treating Swine Waste
Hindawi Publishing Corporation Archaea Volume 2016, Article ID 4089684, 10 pages http://dx.doi.org/10.1155/2016/4089684 Research Article Archaea and Bacteria Acclimate to High Total Ammonia in a Methanogenic Reactor Treating Swine Waste Sofia Esquivel-Elizondo,1,2 Prathap Parameswaran,3 Anca G. Delgado,1 Juan Maldonado,1 Bruce E. Rittmann,1,2 and Rosa Krajmalnik-Brown1,2 1 Swette Center for Environmental Biotechnology, The Biodesign Institute, Arizona State University, P.O. Box 875701, Tempe, AZ 85287-5701, USA 2School of Sustainable Engineering and the Built Environment, Arizona State University, Tempe, AZ, USA 3Department of Civil Engineering, Kansas State University, 2118 Fiedler Hall, Manhattan, KS 66506, USA Correspondence should be addressed to Rosa Krajmalnik-Brown; [email protected] Received 10 June 2016; Accepted 11 August 2016 Academic Editor: Jessica A. Smith Copyright © 2016 Sofia Esquivel-Elizondo et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Inhibition by ammonium at concentrations above 1000 mgN/L is known to harm the methanogenesis phase of anaerobic digestion. We anaerobically digested swine waste and achieved steady state COD-removal efficiency of around 52% with no fatty-acid or H2 accumulation. As the anaerobic microbial community adapted to the gradual increase of total ammonia-N (NH3-N) from 890 ± 295 to 2040 ± 30 mg/L, the Bacterial and Archaeal communities became less diverse. Phylotypes most closely related to hydrogenotrophic Methanoculleus (36.4%) and Methanobrevibacter (11.6%), along with acetoclastic Methanosaeta (29.3%), became the most abundant Archaeal sequences during acclimation. -

Comparative Genomics of Syntrophic Branched-Chain Fatty Acid Degrading Bacteria
Microbes Environ. Vol. 31, No. 3, 288-292, 2016 https://www.jstage.jst.go.jp/browse/jsme2 doi:10.1264/jsme2.ME16057 Comparative Genomics of Syntrophic Branched-Chain Fatty Acid Degrading Bacteria TAKASHI NARIHIRO1,2†*, MASARU K. NOBU2†, HIDEYUKI TAMAKI1,3, YOICHI KAMAGATA1, YUJI SEKIGUCHI4, and WEN-TSO LIU2 1Bioproduction Research Institute, National Institute of Advanced Industrial Science and Technology (AIST), Central 6, Higashi 1–1–1, Tsukuba, Ibaraki 305–8566, Japan; 2Department of Civil and Environmental Engineering, University of Illinois at Urbana-Champaign, 205 North Mathews Ave, Urbana, IL 61801, USA; 3Biotechnology Research Institute, The University of Tokyo, 1–1–1 Yayoi, Bunkyo-ku, Tokyo 113–8657, Japan; and 4Biomedical Research Institute, National Institute of Advanced Industrial Science and Technology (AIST), Central 6, Higashi 1–1–1, Tsukuba, Ibaraki 305–8566, Japan (Received March 16, 2016—Accepted June 5, 2016—Published online July 16, 2016) The syntrophic degradation of branched-chain fatty acids (BCFAs) such as 2-methylbutyrate and isobutyrate is an essential step in the production of methane from proteins/amino acids in anaerobic ecosystems. While a few syntrophic BCFA-degrading bacteria have been isolated, their metabolic pathways in BCFA and short-chain fatty acid (SCFA) degradation as well as energy conservation systems remain unclear. In an attempt to identify these pathways, we herein performed comparative genomics of three syntrophic bacteria: 2-methylbutyrate-degrading “Syntrophomonas wolfei subsp. methylbutyratica” strain JCM 14075T (=4J5T), isobutyrate-degrading Syntrophothermus lipocalidus strain TGB-C1T, and non-BCFA-metabolizing S. wolfei subsp. wolfei strain GöttingenT. We demonstrated that 4J5 and TGB-C1 both encode multiple genes/gene clusters involved in β-oxidation, as observed in the Göttingen genome, which has multiple copies of genes associated with butyrate degradation.