Peat: Home to Novel Syntrophic Species That Feed Acetate- and Hydrogen-Scavenging Methanogens
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Core Sulphate-Reducing Microorganisms in Metal-Removing Semi-Passive Biochemical Reactors and the Co-Occurrence of Methanogens
microorganisms Article Core Sulphate-Reducing Microorganisms in Metal-Removing Semi-Passive Biochemical Reactors and the Co-Occurrence of Methanogens Maryam Rezadehbashi and Susan A. Baldwin * Chemical and Biological Engineering, University of British Columbia, 2360 East Mall, Vancouver, BC V6T 1Z3, Canada; [email protected] * Correspondence: [email protected]; Tel.: +1-604-822-1973 Received: 2 January 2018; Accepted: 17 February 2018; Published: 23 February 2018 Abstract: Biochemical reactors (BCRs) based on the stimulation of sulphate-reducing microorganisms (SRM) are emerging semi-passive remediation technologies for treatment of mine-influenced water. Their successful removal of metals and sulphate has been proven at the pilot-scale, but little is known about the types of SRM that grow in these systems and whether they are diverse or restricted to particular phylogenetic or taxonomic groups. A phylogenetic study of four established pilot-scale BCRs on three different mine sites compared the diversity of SRM growing in them. The mine sites were geographically distant from each other, nevertheless the BCRs selected for similar SRM types. Clostridia SRM related to Desulfosporosinus spp. known to be tolerant to high concentrations of copper were members of the core microbial community. Members of the SRM family Desulfobacteraceae were dominant, particularly those related to Desulfatirhabdium butyrativorans. Methanogens were dominant archaea and possibly were present at higher relative abundances than SRM in some BCRs. Both hydrogenotrophic and acetoclastic types were present. There were no strong negative or positive co-occurrence correlations of methanogen and SRM taxa. Knowing which SRM inhabit successfully operating BCRs allows practitioners to target these phylogenetic groups when selecting inoculum for future operations. -

Fatty Acid Diets: Regulation of Gut Microbiota Composition and Obesity and Its Related Metabolic Dysbiosis
International Journal of Molecular Sciences Review Fatty Acid Diets: Regulation of Gut Microbiota Composition and Obesity and Its Related Metabolic Dysbiosis David Johane Machate 1, Priscila Silva Figueiredo 2 , Gabriela Marcelino 2 , Rita de Cássia Avellaneda Guimarães 2,*, Priscila Aiko Hiane 2 , Danielle Bogo 2, Verônica Assalin Zorgetto Pinheiro 2, Lincoln Carlos Silva de Oliveira 3 and Arnildo Pott 1 1 Graduate Program in Biotechnology and Biodiversity in the Central-West Region of Brazil, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; [email protected] (D.J.M.); [email protected] (A.P.) 2 Graduate Program in Health and Development in the Central-West Region of Brazil, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; pri.fi[email protected] (P.S.F.); [email protected] (G.M.); [email protected] (P.A.H.); [email protected] (D.B.); [email protected] (V.A.Z.P.) 3 Chemistry Institute, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; [email protected] * Correspondence: [email protected]; Tel.: +55-67-3345-7416 Received: 9 March 2020; Accepted: 27 March 2020; Published: 8 June 2020 Abstract: Long-term high-fat dietary intake plays a crucial role in the composition of gut microbiota in animal models and human subjects, which affect directly short-chain fatty acid (SCFA) production and host health. This review aims to highlight the interplay of fatty acid (FA) intake and gut microbiota composition and its interaction with hosts in health promotion and obesity prevention and its related metabolic dysbiosis. -

From Sporulation to Intracellular Offspring Production: Evolution
FROM SPORULATION TO INTRACELLULAR OFFSPRING PRODUCTION: EVOLUTION OF THE DEVELOPMENTAL PROGRAM OF EPULOPISCIUM A Dissertation Presented to the Faculty of the Graduate School of Cornell University In Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy by David Alan Miller January 2012 © 2012 David Alan Miller FROM SPORULATION TO INTRACELLULAR OFFSPRING PRODUCTION: EVOLUTION OF THE DEVELOPMENTAL PROGRAM OF EPULOPISCIUM David Alan Miller, Ph. D. Cornell University 2012 Epulopiscium sp. type B is an unusually large intestinal symbiont of the surgeonfish Naso tonganus. Unlike most other bacteria, Epulopiscium sp. type B has never been observed to undergo binary fission. Instead, to reproduce, it forms multiple intracellular offspring. We believe this process is related to endospore formation, an ancient and complex developmental process performed by certain members of the Firmicutes. Endospore formation has been studied for over 50 years and is best characterized in Bacillus subtilis. To study the evolution of endospore formation in the Firmicutes and the relatedness of this process to intracellular offspring formation in Epulopiscium, we have searched for sporulation genes from the B. subtilis model in all of the completed genomes of members of the Firmicutes, in addition to Epulopiscium sp. type B and its closest relative, the spore-forming Cellulosilyticum lentocellum. By determining the presence or absence of spore genes, we see the evolution of endospore formation in closely related bacteria within the Firmicutes and begin to predict if 19 previously characterized non-spore-formers have the genetic capacity to form a spore. We can also map out sporulation-specific mechanisms likely being used by Epulopiscium for offspring formation. -

Discovery of a Novel Methanogen Prevalent in Thawing Permafrost
ARTICLE Received 11 Jun 2013 | Accepted 7 Jan 2014 | Published 14 Feb 2014 DOI: 10.1038/ncomms4212 Discovery of a novel methanogen prevalent in thawing permafrost Rhiannon Mondav1,*,w, Ben J. Woodcroft1,*, Eun-Hae Kim2, Carmody K. McCalley3,w, Suzanne B. Hodgkins4, Patrick M. Crill5, Jeffrey Chanton4, Gregory B. Hurst6, Nathan C. VerBerkmoes6,w, Scott R. Saleska3, Philip Hugenholtz1, Virginia I. Rich2 & Gene W. Tyson1 Thawing permafrost promotes microbial degradation of cryo-sequestered and new carbon leading to the biogenic production of methane, creating a positive feedback to climate change. Here we determine microbial community composition along a permafrost thaw gradient in northern Sweden. Partially thawed sites were frequently dominated by a single archaeal phylotype, Candidatus ‘Methanoflorens stordalenmirensis’ gen. nov. sp. nov., belonging to the uncultivated lineage ‘Rice Cluster II’ (Candidatus ‘Methanoflorentaceae’ fam. nov.). Metage- nomic sequencing led to the recovery of its near-complete genome, revealing the genes necessary for hydrogenotrophic methanogenesis. These genes are highly expressed and methane carbon isotope data are consistent with hydrogenotrophic production of methane in the partially thawed site. In addition to permafrost wetlands, ‘Methanoflorentaceae’ are widespread in high methane-flux habitats suggesting that this lineage is both prevalent and a major contributor to global methane production. In thawing permafrost, Candidatus ‘M. stordalenmirensis’ appears to be a key mediator of methane-based positive feedback to climate warming. 1 Australian Centre for Ecogenomics, School of Chemistry and Molecular Biosciences, University of Queensland, Brisbane 4072, Queensland, Australia. 2 Department of Soil, Water and Environmental Science, University of Arizona, Tucson, Arizona 85721, USA. 3 Ecology and Evolutionary Biology Department, University of Arizona, Tucson, Arizona 85721, USA. -
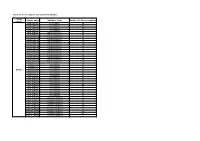
Table S1: List of Samples Included in the Analysis
Table S1: list of samples included in the analysis Study Sample name Inhibitory status Number of days at sampling number DNA.0P2T4 No inhibition 29 DNA.0P2T6 No inhibition 57 DNA.10P2T4 No inhibition 29 DNA.10P2T6 No inhibition 57 DNA.75P2T4 Phenol inhibition 29 DNA.75P2T6 Phenol inhibition 57 DNA.100P2T4 Phenol inhibition 29 DNA.100P2T6 Phenol inhibition 57 DNA.125P1T4 Phenol inhibition 29 DNA.125P1T6 Phenol inhibition 57 DNA.125P2T4 Phenol inhibition 29 DNA.125P2T6 Phenol inhibition 57 DNA.125P3T4 Phenol inhibition 29 DNA.125P3T6 Phenol inhibition 57 DNA.150P2T4 Phenol inhibition 29 DNA.150P2T6 Phenol inhibition 57 DNA.200P2T4 Phenol inhibition 29 DNA.200P2T6 Phenol inhibition 57 DNA.0N2T4 No inhibition 29 DNA.0N2T5 No inhibition 42 DNA.0N2T6 No inhibition 57 Study 1 DNA.5N2T4 No inhibition 29 DNA.5N2T5 No inhibition 42 DNA.5N2T6 No inhibition 57 DNA.10N2T4 No inhibition 29 DNA.10N2T5 No inhibition 42 DNA.10N2T6 No inhibition 57 DNA.15N2T4 No inhibition 29 DNA.15N2T5 No inhibition 42 DNA.15N2T6 No inhibition 57 DNA.25N2T4 No inhibition 29 DNA.25N2T5 No inhibition 42 DNA.25N2T6 No inhibition 57 DNA.75N2T4 Ammonia inhibition 29 DNA.75N2T5 Ammonia inhibition 42 DNA.75N2T6 Ammonia inhibition 57 DNA.100N2T4 Ammonia inhibition 29 DNA.100N2T5 Ammonia inhibition 42 DNA.100N2T6 Ammonia inhibition 57 DNA.250N2T4 Ammonia inhibition 29 DNA.250N2T5 Ammonia inhibition 42 DNA.250N2T6 Ammonia inhibition 57 nono2T3 No inhibition 16 noN2T4 Ammonia inhibition 23 noN2T8 Ammonia inhibition 60 noN2T9 Ammonia inhibition 85 noPhi2T4 Phenol inhibition 23 noPhi2T5 -

Dialogue on the Nomenclature and Classification of Prokaryotes
G Model SYAPM-25929; No. of Pages 10 ARTICLE IN PRESS Systematic and Applied Microbiology xxx (2018) xxx–xxx Contents lists available at ScienceDirect Systematic and Applied Microbiology journal homepage: www.elsevier.de/syapm Dialogue on the nomenclature and classification of prokaryotes a,∗ b Ramon Rosselló-Móra , William B. Whitman a Marine Microbiology Group, IMEDEA (CSIC-UIB), 07190 Esporles, Spain b Department of Microbiology, University of Georgia, Athens, GA 30602, USA a r t i c l e i n f o a b s t r a c t Keywords: The application of next generation sequencing and molecular ecology to the systematics and taxonomy Bacteriological code of prokaryotes offers enormous insights into prokaryotic biology. This discussion explores some major Taxonomy disagreements but also considers the opportunities associated with the nomenclature of the uncultured Nomenclature taxa, the use of genome sequences as type material, the plurality of the nomenclatural code, and the roles Candidatus of an official or computer-assisted taxonomy. Type material © 2018 Elsevier GmbH. All rights reserved. Is naming important when defined here as providing labels Prior to the 1980s, one of the major functions of Bergey’s Manual for biological entities? was to associate the multiple names in current usage with the cor- rect organism. For instance, in the 1948 edition of the Manual, 21 Whitman and 33 names were associated with the common bacterial species now named Escherichia coli and Bacillus subtilis, respectively [5]. When discussing the nomenclature of prokaryotes, we must first This experience illustrates that without a naming system gener- establish the role and importance of naming. -

Pan-Genome Analysis and Ancestral State Reconstruction Of
www.nature.com/scientificreports OPEN Pan‑genome analysis and ancestral state reconstruction of class halobacteria: probability of a new super‑order Sonam Gaba1,2, Abha Kumari2, Marnix Medema 3 & Rajeev Kaushik1* Halobacteria, a class of Euryarchaeota are extremely halophilic archaea that can adapt to a wide range of salt concentration generally from 10% NaCl to saturated salt concentration of 32% NaCl. It consists of the orders: Halobacteriales, Haloferaciales and Natriabales. Pan‑genome analysis of class Halobacteria was done to explore the core (300) and variable components (Softcore: 998, Cloud:36531, Shell:11784). The core component revealed genes of replication, transcription, translation and repair, whereas the variable component had a major portion of environmental information processing. The pan‑gene matrix was mapped onto the core‑gene tree to fnd the ancestral (44.8%) and derived genes (55.1%) of the Last Common Ancestor of Halobacteria. A High percentage of derived genes along with presence of transformation and conjugation genes indicate the occurrence of horizontal gene transfer during the evolution of Halobacteria. A Core and pan‑gene tree were also constructed to infer a phylogeny which implicated on the new super‑order comprising of Natrialbales and Halobacteriales. Halobacteria1,2 is a class of phylum Euryarchaeota3 consisting of extremely halophilic archaea found till date and contains three orders namely Halobacteriales4,5 Haloferacales5 and Natrialbales5. Tese microorganisms are able to dwell at wide range of salt concentration generally from 10% NaCl to saturated salt concentration of 32% NaCl6. Halobacteria, as the name suggests were once considered a part of a domain "Bacteria" but with the discovery of the third domain "Archaea" by Carl Woese et al.7, it became part of Archaea. -

1 Structure and Function of the Methanogenic Microbial Communities in Soils from 1 Flooded Rice and Upland Soybean Fields from S
*Manuscript with continous line numbering Click here to view linked References 1 Structure and function of the methanogenic microbial communities in soils from 2 flooded rice and upland soybean fields from Sanjiang Plain, NE China 3 4 Marcela Hernándeza,c*, Ralf Conrada, Melanie Klosea, Ke Mab, Yahai Lub 5 6 a Max Planck Institute for Terrestrial Microbiology, Karl-von-Frisch-Str. 10, 35043 7 Marburg, Germany 8 b College of Resources and Environmental Sciences, China Agricultural University, Beijing, 9 China 10 c Centre for Biological Sciences, University of Southampton, Southampton, SO17 1BJ 11 United Kingdom 12 13 14 15 16 17 18 *Corresponding author. Marcela Hernández: Centre for Biological Sciences, University of 19 Southampton, Southampton, SO17 1BJ United Kingdom. Tel.: +44 (0) 23 8059 4291. E- 20 mail address: [email protected] 21 1 22 Abstract 23 About 50 years ago, most of the natural wetlands in northeast China, the Sanjiang plain, 24 were converted to either flooded rice fields or to upland soybean fields. After the 25 conversion, natural wetland soils were either managed as artificial wetland or as drained 26 upland resulting in soil microbial community changes. The purpose of our study was to 27 understand how methanogenic microbial communities and their functions had changed in 28 the two different soils upon conversion, and whether these communities now exhibit 29 different resistance/resilience to drying and rewetting. Therefore, we determined function, 30 abundance and composition of the methanogenic archaeal and bacterial communities in two 31 soils reclaimed from a Carex wetland 25 years ago. -

Desulfovirga Adipica Gen. Nov., Sp. Nov., an Adipate-Degrading, Gram-Negative, Sulfate-Reducing Bacterium
International Journal of Systematic and Evolutionary Microbiology (2000), 50, 639–644 Printed in Great Britain Desulfovirga adipica gen. nov., sp. nov., an adipate-degrading, Gram-negative, sulfate-reducing bacterium Kazuhiro Tanaka,1 Erko Stackebrandt,2 Shigehiro Tohyama3 and Tadashi Eguchi3 Author for correspondence: Kazuhiro Tanaka. Tel\Fax: j81 298 61 6083. e-mail: ktanaka!nibh.go.jp 1 Applied Microbiology A novel, mesophilic, Gram-negative bacterium was isolated from an anaerobic Department, National digestor for municipal wastewater. The bacterium degraded adipate in the Institute of Bioscience and Human-Technology, presence of sulfate, sulfite, thiosulfate and elemental sulfur. (E)-2- Higashi 1-1, Tsukuba, Hexenedioate accumulated transiently in the degradation of adipate. (E)-2- Ibaraki 305-8566, Japan Hexenedioate, (E)-3-hexenedioate, pyruvate, lactate, C1–C12 straight-chain fatty 2 Deutsche Sammlung von acids and C2–C10 straight-chain primary alcohols were also utilized as electron Mikroorganismen und donors. 3-Phenylpropionate was oxidized to benzoate. The GMC content of the Zellkulturen GmbH, Mascheroder Weg 1b, DNA was 60 mol%. 16S rDNA sequence analysis revealed that the new isolate D-38124 Braunschweig, clustered with species of the genus Syntrophobacter and Desulforhabdus Germany amnigenus. Strain TsuAS1T resembles Desulforhabdus amnigenus DSM 10338T 3 Department of Chemistry with respect to the ability to utilize acetate as an electron donor and the and Materials Science, inability to utilize propionate without sulfate in co-culture with Tokyo Institute of T T Technology, O-okayama, Methanospirillum hungatei DSM 864. Strains TsuAS1 and DSM 10338 form a Meguro-ku, Tokyo ‘non-syntrophic subcluster’ within the genus Syntrophobacter. Desulfovirga 152-8551, Japan adipica gen. -

Sp. Nov., New Syntrophically Propionate-Oxidizing Anaerobe Growing in Pure Culture with Propionate and Sulfate
Arch Microbiol (1995) 164:346-352 Springer-Verlag 1995 Christina Wallrabenstein Elisabeth Hauschild Bernhard Schink Syntrophobacter pfennigii sp. nov., new syntrophically propionate-oxidizing anaerobe growing in pure culture with propionate and sulfate Received: 3 July 1995 / Accepted: 16 August 1995 Abstract A new strain of syntrophically propionate-oxi- teria (Zehnder 1978). Fermentation of propionate to ac- dizing fermenting bacteria, strain KoPropl, was isolated etate, CO2, and hydrogen is a highly endergonic process from anoxic sludge of a municipal sewage plant. It oxi- (calculations of free energies after Thauer et al. 1977): dized propionate or lactate in cooperation with the hydro- CH3CH2COO + 2 H20----)CH3COO -t-CO2 + 3 H 2 (1) gen- and formate-utilizing Methanospirillum hungatei AG 0" = +76.0 kJ/mol propionate and grew as well in pure culture without a syntrophic part- ner with propionate or lactate plus sulfate as energy The hydrogen partial pressure has to be kept low by the source. In all cases, the substrates were oxidized stoichio- partner organism to make the reaction energetically feasi- metrically to acetate and CO2, with concomitant forma- ble, e.g., in syntrophic methanogenic propionate degrada- tion of methane or sulfide. Cells formed gas vesicles in tion: the late growth phase and contained cytochromes b and c, 4 CH3CH2COO- + 2 H20--M CH3COO- + CO2+3 CH 4 (2) a menaquinone-7, and desulforubidin, but no desul- AG 0" = -26.5 kJ/mol propionate foviridin. Enzyme measurements in cell-free extracts indi- cated that propionate was oxidized through the methyl- However, the amount of free energy liberated during syn- malonyl CoA pathway. -

'Candidatus Desulfonatronobulbus Propionicus': a First Haloalkaliphilic
Delft University of Technology ‘Candidatus Desulfonatronobulbus propionicus’ a first haloalkaliphilic member of the order Syntrophobacterales from soda lakes Sorokin, D. Y.; Chernyh, N. A. DOI 10.1007/s00792-016-0881-3 Publication date 2016 Document Version Accepted author manuscript Published in Extremophiles: life under extreme conditions Citation (APA) Sorokin, D. Y., & Chernyh, N. A. (2016). ‘Candidatus Desulfonatronobulbus propionicus’: a first haloalkaliphilic member of the order Syntrophobacterales from soda lakes. Extremophiles: life under extreme conditions, 20(6), 895-901. https://doi.org/10.1007/s00792-016-0881-3 Important note To cite this publication, please use the final published version (if applicable). Please check the document version above. Copyright Other than for strictly personal use, it is not permitted to download, forward or distribute the text or part of it, without the consent of the author(s) and/or copyright holder(s), unless the work is under an open content license such as Creative Commons. Takedown policy Please contact us and provide details if you believe this document breaches copyrights. We will remove access to the work immediately and investigate your claim. This work is downloaded from Delft University of Technology. For technical reasons the number of authors shown on this cover page is limited to a maximum of 10. Extremophiles DOI 10.1007/s00792-016-0881-3 ORIGINAL PAPER ‘Candidatus Desulfonatronobulbus propionicus’: a first haloalkaliphilic member of the order Syntrophobacterales from soda lakes D. Y. Sorokin1,2 · N. A. Chernyh1 Received: 23 August 2016 / Accepted: 4 October 2016 © Springer Japan 2016 Abstract Propionate can be directly oxidized anaerobi- from its members at the genus level. -

Diversity of Methane-Cycling Microorganisms in Soils and Their Relation to Oxygen
Diversity of Methane-cycling Microorganisms in Soils and Their Relation to Oxygen Claudia Knief* Institute of Crop Science and Resource Conservation – Molecular Biology of the Rhizosphere, University of Bonn, Bonn, Germany. *Correspondence: [email protected] htps://doi.org/10.21775/cimb.033.023 Abstract Introduction Microorganisms are important players in the Methane cycling microorganisms are of interest global methane cycle. Anaerobic methanogenic for microbiologists since more than a century. archaea are largely responsible for methane pro- Research on these microorganisms was initially duction, while aerobic methanotrophic bacteria, largely driven by the curiosity to understand their as well as anaerobic methanotrophic bacteria particular physiology that leads to the production and archaea, are involved in methane oxidation. or consumption of methane. While this interest is In anoxic wetland soils, methanogens produce still a driver, the importance of methane as green- methane, while methanotrophs act as a flter house gas has become another important factor, and reduce methane emissions. In the predomi- promoting further research on methanogenic and nantly oxic upland soils, aerobic methanotrophs methanotrophic microorganisms. Tis leads to a oxidize atmospheric methane. Tis review gives continuously beter understanding of their physi- an overview of the diversity of methanogenic ology and ecology, and it becomes evident that and methanotrophic microorganisms, highlights the processes of microbial methane production recent discoveries and provides information and consumption are mediated by more com- concerning their occurrence in soils. Recent plex functional guilds than initially thought. Te fndings indicate that the methanogenic and improved understanding is not only due to the con- methanotrophic lifestyles are more widespread stantly increasing diversity of methanogenic and in microorganisms than previously thought, and methanotrophic microorganisms (e.g.