DNA-SIP Based Genome-Centric Metagenomics Identifies Key Long-Chain Fatty Acid-Degrading Populations in Anaerobic Digesters with Different Feeding Frequencies
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Methane Production on Rock and Soil Substrates by Methanogens: Implications for Life on Mars
Bioastronomy 2007: Molecules, Microbes, and Extraterrestrial Life ASP Conference Series, Vol. 420, 2009 K. J. Meech, J. V. Keane, M. J. Mumma, J. L. Siefert, and D. J. Werthimer, eds. Methane Production on Rock and Soil Substrates by Methanogens: Implications for Life on Mars H. A. Kozup and T. A. Kral 1West Virginia University, WV 26505 2Arkansas Center for Space and Planetary Sciences, University of Arkansas, Fayetteville 72701 Abstract. In order to understand the methanogens as models for possible life on Mars, and some of the factors likely to be important in determining their abundance and distribution, we have measured their ability to produce methane on a few types of inorganic rock and soil substrates. Since organic ma- terials have not been detected in measurable quantities at the surface of Mars, there is no reason to believe that they would exist in the subsurface. Samples of three methanogens (Methanosarcina barkeri, Methanobacterium formicicum, and Methanothermobacter wolfeii) were placed on four substrates (sand, gravel, basalt, and a Mars soil simulant, JSC Mars-1) and methane production mea- sured. Glass beads were used as a control substrate. As in earlier experiments with JSC Mars-1 soil simulant, a crushed volcanic tephra, methane was pro- duced by all three methanogens when placed on the substrates, sand and gravel. None produced methane on basalt in these experiments, a mineral common in Martian soil. While these substrates do not represent the full range of materials likely to be present on the surface of Mars, the present results suggest that while some surface materials on Mars may not support this type of organism, others might. -

Fatty Acid Diets: Regulation of Gut Microbiota Composition and Obesity and Its Related Metabolic Dysbiosis
International Journal of Molecular Sciences Review Fatty Acid Diets: Regulation of Gut Microbiota Composition and Obesity and Its Related Metabolic Dysbiosis David Johane Machate 1, Priscila Silva Figueiredo 2 , Gabriela Marcelino 2 , Rita de Cássia Avellaneda Guimarães 2,*, Priscila Aiko Hiane 2 , Danielle Bogo 2, Verônica Assalin Zorgetto Pinheiro 2, Lincoln Carlos Silva de Oliveira 3 and Arnildo Pott 1 1 Graduate Program in Biotechnology and Biodiversity in the Central-West Region of Brazil, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; [email protected] (D.J.M.); [email protected] (A.P.) 2 Graduate Program in Health and Development in the Central-West Region of Brazil, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; pri.fi[email protected] (P.S.F.); [email protected] (G.M.); [email protected] (P.A.H.); [email protected] (D.B.); [email protected] (V.A.Z.P.) 3 Chemistry Institute, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; [email protected] * Correspondence: [email protected]; Tel.: +55-67-3345-7416 Received: 9 March 2020; Accepted: 27 March 2020; Published: 8 June 2020 Abstract: Long-term high-fat dietary intake plays a crucial role in the composition of gut microbiota in animal models and human subjects, which affect directly short-chain fatty acid (SCFA) production and host health. This review aims to highlight the interplay of fatty acid (FA) intake and gut microbiota composition and its interaction with hosts in health promotion and obesity prevention and its related metabolic dysbiosis. -

Histone Variants in Archaea and the Evolution of Combinatorial Chromatin Complexity
Histone variants in archaea and the evolution of combinatorial chromatin complexity Kathryn M. Stevensa,b, Jacob B. Swadlinga,b, Antoine Hochera,b, Corinna Bangc,d, Simonetta Gribaldoe, Ruth A. Schmitzc, and Tobias Warneckea,b,1 aMolecular Systems Group, Quantitative Biology Section, Medical Research Council London Institute of Medical Sciences, London W12 0NN, United Kingdom; bInstitute of Clinical Sciences, Faculty of Medicine, Imperial College London, London W12 0NN, United Kingdom; cInstitute for General Microbiology, University of Kiel, 24118 Kiel, Germany; dInstitute of Clinical Molecular Biology, University of Kiel, 24105 Kiel, Germany; and eDepartment of Microbiology, Unit “Evolutionary Biology of the Microbial Cell,” Institut Pasteur, 75015 Paris, France Edited by W. Ford Doolittle, Dalhousie University, Halifax, NS, Canada, and approved October 28, 2020 (received for review April 14, 2020) Nucleosomes in eukaryotes act as platforms for the dynamic inte- additional histone dimers can be taggedontothistetramertoyield gration of epigenetic information. Posttranslational modifications oligomers of increasing length that wrap correspondingly more DNA are reversibly added or removed and core histones exchanged for (3, 6–9). Almost all archaeal histones lack tails and PTMs have yet to paralogous variants, in concert with changing demands on tran- be reported. Many archaea do, however, encode multiple histone scription and genome accessibility. Histones are also common in paralogs (8, 10) that can flexibly homo- and heterodimerize in -

From Sporulation to Intracellular Offspring Production: Evolution
FROM SPORULATION TO INTRACELLULAR OFFSPRING PRODUCTION: EVOLUTION OF THE DEVELOPMENTAL PROGRAM OF EPULOPISCIUM A Dissertation Presented to the Faculty of the Graduate School of Cornell University In Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy by David Alan Miller January 2012 © 2012 David Alan Miller FROM SPORULATION TO INTRACELLULAR OFFSPRING PRODUCTION: EVOLUTION OF THE DEVELOPMENTAL PROGRAM OF EPULOPISCIUM David Alan Miller, Ph. D. Cornell University 2012 Epulopiscium sp. type B is an unusually large intestinal symbiont of the surgeonfish Naso tonganus. Unlike most other bacteria, Epulopiscium sp. type B has never been observed to undergo binary fission. Instead, to reproduce, it forms multiple intracellular offspring. We believe this process is related to endospore formation, an ancient and complex developmental process performed by certain members of the Firmicutes. Endospore formation has been studied for over 50 years and is best characterized in Bacillus subtilis. To study the evolution of endospore formation in the Firmicutes and the relatedness of this process to intracellular offspring formation in Epulopiscium, we have searched for sporulation genes from the B. subtilis model in all of the completed genomes of members of the Firmicutes, in addition to Epulopiscium sp. type B and its closest relative, the spore-forming Cellulosilyticum lentocellum. By determining the presence or absence of spore genes, we see the evolution of endospore formation in closely related bacteria within the Firmicutes and begin to predict if 19 previously characterized non-spore-formers have the genetic capacity to form a spore. We can also map out sporulation-specific mechanisms likely being used by Epulopiscium for offspring formation. -

Crystalline Iron Oxides Stimulate Methanogenic Benzoate Degradation in Marine Sediment-Derived Enrichment Cultures
The ISME Journal https://doi.org/10.1038/s41396-020-00824-7 ARTICLE Crystalline iron oxides stimulate methanogenic benzoate degradation in marine sediment-derived enrichment cultures 1,2 1 3 1,2 1 David A. Aromokeye ● Oluwatobi E. Oni ● Jan Tebben ● Xiuran Yin ● Tim Richter-Heitmann ● 2,4 1 5 5 1 2,3 Jenny Wendt ● Rolf Nimzyk ● Sten Littmann ● Daniela Tienken ● Ajinkya C. Kulkarni ● Susann Henkel ● 2,4 2,4 1,3 2,3,4 1,2 Kai-Uwe Hinrichs ● Marcus Elvert ● Tilmann Harder ● Sabine Kasten ● Michael W. Friedrich Received: 19 May 2020 / Revised: 9 October 2020 / Accepted: 22 October 2020 © The Author(s) 2020. This article is published with open access Abstract Elevated dissolved iron concentrations in the methanic zone are typical geochemical signatures of rapidly accumulating marine sediments. These sediments are often characterized by co-burial of iron oxides with recalcitrant aromatic organic matter of terrigenous origin. Thus far, iron oxides are predicted to either impede organic matter degradation, aiding its preservation, or identified to enhance organic carbon oxidation via direct electron transfer. Here, we investigated the effect of various iron oxide phases with differing crystallinity (magnetite, hematite, and lepidocrocite) during microbial 1234567890();,: 1234567890();,: degradation of the aromatic model compound benzoate in methanic sediments. In slurry incubations with magnetite or hematite, concurrent iron reduction, and methanogenesis were stimulated during accelerated benzoate degradation with methanogenesis as the dominant electron sink. In contrast, with lepidocrocite, benzoate degradation, and methanogenesis were inhibited. These observations were reproducible in sediment-free enrichments, even after five successive transfers. Genes involved in the complete degradation of benzoate were identified in multiple metagenome assembled genomes. -

Methanobacterium Paludis Sp. Nov. and a Novel Strain of Methanobacterium Lacus Isolated from Northern Peatlands
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by The University of North Carolina at Greensboro Archived version from NCDOCKS Institutional Repository http://libres.uncg.edu/ir/asu/ Methanobacterium Paludis Sp. Nov. And A Novel Strain Of Methanobacterium Lacus Isolated From Northern Peatlands By: Suzanna L. Brauer, Hinsby Cadillo-Quiroz, Noah Goodson, Joseph B. Yavitt & Stephen H. Zinder Abstract Two mesophilic, hydrogenotrophic methanogens, designated strains SWAN1T and AL-21, were isolated from two contrasting peatlands: a near circumneutral temperate minerotrophic fen in New York State, USA, and an acidic boreal poor fen site in Alaska, USA, respectively. Cells of the two strains were rod-shaped, non- motile, stained Gram-negative and resisted lysis with 0.1 % SDS. Cell size was 0.6 1.5–2.8 mm for strain SWAN1T and 0.45–0.85 1.5–35 mm for strain AL-21. The strains used H2/CO2 but not formate or other substrates for methanogenesis, grew optimally around 32–37 6C, and their growth spanned through a slightly low to neutral pH range (4.7–7.1). Strain AL-21 grew optimally closer to neutrality at pH 6.2, whereas strain SWAN1T showed a lower optimal pH at 5.4–5.7. The two strains were sensitive to NaCl with a maximal tolerance at 160 mM for strain SWAN1T and 50 mM for strain AL-21. Na2S was toxic at very low concentrations (0.01–0.8 mM), resulting in growth inhibition above these values. The DNA G+C content of the genomes was 35.7 mol% for strain SWAN1T and 35.8 mol% for strain AL-21. -

METHANOGENS AS MODELS for LIFE on MARS. R. L. Mickol1, W. H. Waddell2, and T
Eighth International Conference on Mars (2014) 1005.pdf METHANOGENS AS MODELS FOR LIFE ON MARS. R. L. Mickol1, W. H. Waddell2, and T. A. Kral1,3, 1Arkansas Center for Space and Planetary Sciences, 202 Old Museum Building, University of Arkansas, Fayetteville, Arkansas, 72701, USA, [[email protected]], 2Dept. of Health, Human Performance, and Recreation, HPER 308, University of Arkansas, Fayetteville, Arkansas, 72701, USA, 3Dept. of Biological Sciences, SCEN 632, University of Arkansas, Fayetteville, Arkansas, 72701, USA. Introduction: The discovery of methane in the Sciences, University of Arkansas, Fayetteville, Arkan- martian atmosphere [1-4] has fueled the study of meth- sas. All four methanogen species were tested in these anogens as ideal candidates for life on Mars. Methano- experiments. Methanogens were grown in their respec- gens are chemoautotrophs from the domain Archaea. tive anaerobic growth media and placed into the cham- These microorganisms utilize hydrogen as an energy ber with a palladium catalyst box to remove residual source and carbon dioxide as a carbon source to pro- oxygen. The chamber was evacuated to a pre- duce methane. Methanogens can be considered ideal determined pressure and filled with 80:20 H2:CO2 gas. candidates for life on Mars because they are anaerobic, This procedure was repeated three times to ensure re- they do not require organic nutrients and are non- moval of the atmosphere. The chamber was then main- photosynthetic, indicating they could exist in sub- tained at the desired pressure (133-143 mbar, 67-72 surface environments. mbar, 33-38 mbar, 6-10 mbar, 7-20 mbar) for the dura- Our lab has studied methanogens as models for life tion of the experiments. -
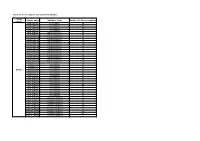
Table S1: List of Samples Included in the Analysis
Table S1: list of samples included in the analysis Study Sample name Inhibitory status Number of days at sampling number DNA.0P2T4 No inhibition 29 DNA.0P2T6 No inhibition 57 DNA.10P2T4 No inhibition 29 DNA.10P2T6 No inhibition 57 DNA.75P2T4 Phenol inhibition 29 DNA.75P2T6 Phenol inhibition 57 DNA.100P2T4 Phenol inhibition 29 DNA.100P2T6 Phenol inhibition 57 DNA.125P1T4 Phenol inhibition 29 DNA.125P1T6 Phenol inhibition 57 DNA.125P2T4 Phenol inhibition 29 DNA.125P2T6 Phenol inhibition 57 DNA.125P3T4 Phenol inhibition 29 DNA.125P3T6 Phenol inhibition 57 DNA.150P2T4 Phenol inhibition 29 DNA.150P2T6 Phenol inhibition 57 DNA.200P2T4 Phenol inhibition 29 DNA.200P2T6 Phenol inhibition 57 DNA.0N2T4 No inhibition 29 DNA.0N2T5 No inhibition 42 DNA.0N2T6 No inhibition 57 Study 1 DNA.5N2T4 No inhibition 29 DNA.5N2T5 No inhibition 42 DNA.5N2T6 No inhibition 57 DNA.10N2T4 No inhibition 29 DNA.10N2T5 No inhibition 42 DNA.10N2T6 No inhibition 57 DNA.15N2T4 No inhibition 29 DNA.15N2T5 No inhibition 42 DNA.15N2T6 No inhibition 57 DNA.25N2T4 No inhibition 29 DNA.25N2T5 No inhibition 42 DNA.25N2T6 No inhibition 57 DNA.75N2T4 Ammonia inhibition 29 DNA.75N2T5 Ammonia inhibition 42 DNA.75N2T6 Ammonia inhibition 57 DNA.100N2T4 Ammonia inhibition 29 DNA.100N2T5 Ammonia inhibition 42 DNA.100N2T6 Ammonia inhibition 57 DNA.250N2T4 Ammonia inhibition 29 DNA.250N2T5 Ammonia inhibition 42 DNA.250N2T6 Ammonia inhibition 57 nono2T3 No inhibition 16 noN2T4 Ammonia inhibition 23 noN2T8 Ammonia inhibition 60 noN2T9 Ammonia inhibition 85 noPhi2T4 Phenol inhibition 23 noPhi2T5 -

EXPERIMENTAL STUDIES on FERMENTATIVE FIRMICUTES from ANOXIC ENVIRONMENTS: ISOLATION, EVOLUTION, and THEIR GEOCHEMICAL IMPACTS By
EXPERIMENTAL STUDIES ON FERMENTATIVE FIRMICUTES FROM ANOXIC ENVIRONMENTS: ISOLATION, EVOLUTION, AND THEIR GEOCHEMICAL IMPACTS By JESSICA KEE EUN CHOI A dissertation submitted to the School of Graduate Studies Rutgers, The State University of New Jersey In partial fulfillment of the requirements For the degree of Doctor of Philosophy Graduate Program in Microbial Biology Written under the direction of Nathan Yee And approved by _______________________________________________________ _______________________________________________________ _______________________________________________________ _______________________________________________________ New Brunswick, New Jersey October 2017 ABSTRACT OF THE DISSERTATION Experimental studies on fermentative Firmicutes from anoxic environments: isolation, evolution and their geochemical impacts by JESSICA KEE EUN CHOI Dissertation director: Nathan Yee Fermentative microorganisms from the bacterial phylum Firmicutes are quite ubiquitous in subsurface environments and play an important biogeochemical role. For instance, fermenters have the ability to take complex molecules and break them into simpler compounds that serve as growth substrates for other organisms. The research presented here focuses on two groups of fermentative Firmicutes, one from the genus Clostridium and the other from the class Negativicutes. Clostridium species are well-known fermenters. Laboratory studies done so far have also displayed the capability to reduce Fe(III), yet the mechanism of this activity has not been investigated -

Syntrophic Butyrate and Propionate Oxidation Processes 491
Environmental Microbiology Reports (2010) 2(4), 489–499 doi:10.1111/j.1758-2229.2010.00147.x Minireview Syntrophic butyrate and propionate oxidation processes: from genomes to reaction mechanismsemi4_147 489..499 Nicolai Müller,1† Petra Worm,2† Bernhard Schink,1* a cytoplasmic fumarate reductase to drive energy- Alfons J. M. Stams2 and Caroline M. Plugge2 dependent succinate oxidation. Furthermore, we 1Faculty for Biology, University of Konstanz, D-78457 propose that homologues of the Thermotoga mar- Konstanz, Germany. itima bifurcating [FeFe]-hydrogenase are involved 2Laboratory of Microbiology, Wageningen University, in NADH oxidation by S. wolfei and S. fumaroxidans Dreijenplein 10, 6703 HB Wageningen, the Netherlands. to form hydrogen. Summary Introduction In anoxic environments such as swamps, rice fields In anoxic environments such as swamps, rice paddy fields and sludge digestors, syntrophic microbial communi- and intestines of higher animals, methanogenic commu- ties are important for decomposition of organic nities are important for decomposition of organic matter to matter to CO2 and CH4. The most difficult step is the CO2 and CH4 (Schink and Stams, 2006; Mcinerney et al., fermentative degradation of short-chain fatty acids 2008; Stams and Plugge, 2009). Moreover, they are the such as propionate and butyrate. Conversion of these key biocatalysts in anaerobic bioreactors that are used metabolites to acetate, CO2, formate and hydrogen is worldwide to treat industrial wastewaters and solid endergonic under standard conditions and occurs wastes. Different types of anaerobes have specified only if methanogens keep the concentrations of these metabolic functions in the degradation pathway and intermediate products low. Butyrate and propionate depend on metabolite transfer which is called syntrophy degradation pathways include oxidation steps of (Schink and Stams, 2006). -

Large Carbon Isotope Variability During Methanogenesis Under Alkaline Conditions Hannah M
Large carbon isotope variability during methanogenesis under alkaline conditions Hannah M. Millera Nabil Chaudhrya Mark E. Conradb Markus Billb Sebastian H. Kopfa Alexis S. Templetona Abstract 13 High carbon isotope values (δ CCH4 > −40‰) have widely been used as evidence that methane in alkaline rock-hosted fluids was formed abiotically, particularly in serpentinizing systems. However, isotope fractionation during microbial methanogenesis is relatively understudied at high pH. We isolated a hydrogenotrophic Methanobacterium sp. from hyperalkaline subsurface fluids in the Samail ophiolite to assess how carbon and hydrogen isotope values of CH4 varied depending upon pH and carbonate mineral source (NaHCO3 or CaCO3). The hydrogen isotope fractionation αH20/CH4 (1.46–1.66) did not vary across pH. In contrast, the expressed carbon isotope fractionation, αCO2/CH4, ranged from 1.028 to 1.089. Carbon isotope fractionation increased with pH, reaching a maximum 13C depletion of −85‰. However, the 13C depletion significantly diminished at pH ≥ 9 for 13 CaCO3-amended experiments, generating δ CCH4 as high as −28‰. To 13 evaluate the large variability in δ CCH4, we developed a steady-state model to assess how the rates of carbonate dissolution, cellular uptake of CO2 and irreversible CH4 production can affect the net isotope fractionation during 13 methanogenesis. Methanobacterium sp. can produce highly depleted δ CCH4 in simulated alkaline serpentinizing fluids when dissolved inorganic carbon levels are high and methanogenesis rates are slow. However, small carbon isotope fractionation occurs when rates of carbonate dissolution are slower 13 than cellular uptake, leading to relatively high δ CCH4 values (>∼−35‰) that are traditionally interpreted to be purely “abiotic”. -

Microbial Communities Mediating Algal Detritus Turnover Under Anaerobic Conditions
Microbial communities mediating algal detritus turnover under anaerobic conditions Jessica M. Morrison1,*, Chelsea L. Murphy1,*, Kristina Baker1, Richard M. Zamor2, Steve J. Nikolai2, Shawn Wilder3, Mostafa S. Elshahed1 and Noha H. Youssef1 1 Department of Microbiology and Molecular Genetics, Oklahoma State University, Stillwater, OK, USA 2 Grand River Dam Authority, Vinita, OK, USA 3 Department of Integrative Biology, Oklahoma State University, Stillwater, OK, USA * These authors contributed equally to this work. ABSTRACT Background. Algae encompass a wide array of photosynthetic organisms that are ubiquitously distributed in aquatic and terrestrial habitats. Algal species often bloom in aquatic ecosystems, providing a significant autochthonous carbon input to the deeper anoxic layers in stratified water bodies. In addition, various algal species have been touted as promising candidates for anaerobic biogas production from biomass. Surprisingly, in spite of its ecological and economic relevance, the microbial community involved in algal detritus turnover under anaerobic conditions remains largely unexplored. Results. Here, we characterized the microbial communities mediating the degradation of Chlorella vulgaris (Chlorophyta), Chara sp. strain IWP1 (Charophyceae), and kelp Ascophyllum nodosum (phylum Phaeophyceae), using sediments from an anaerobic spring (Zodlteone spring, OK; ZDT), sludge from a secondary digester in a local wastewater treatment plant (Stillwater, OK; WWT), and deeper anoxic layers from a seasonally stratified lake