Acipenser Transmontanus) and Implications for Conservation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Carrier Sekani Tribal Council Aboriginal Interests & Use Study On
Carrier Sekani Tribal Council Aboriginal Interests & Use Study on the Enbridge Gateway Pipeline An Assessment of the Impacts of the Proposed Enbridge Gateway Pipeline on the Carrier Sekani First Nations May 2006 Carrier Sekani Tribal Council i Aboriginal Interests & Use Study on the Proposed Gateway Pipeline ACKNOWLEDGEMENTS The Carrier Sekani Tribal Council Aboriginal Interests & Use Study was carried out under the direction of, and by many members of the Carrier Sekani First Nations. This work was possible because of the many people who have over the years established the written records of the history, territories, and governance of the Carrier Sekani. Without this foundation, this study would have been difficult if not impossible. This study involved many community members in various capacities including: Community Coordinators/Liaisons Ryan Tibbetts, Burns Lake Band Bev Ketlo, Nadleh Whut’en First Nation Sara Sam, Nak’azdli First Nation Rosa McIntosh, Saik’uz First Nation Bev Bird & Ron Winser, Tl’azt’en Nation Michael Teegee & Terry Teegee, Takla Lake First Nation Viola Turner, Wet’suwet’en First Nation Elders, Trapline & Keyoh Holders Interviewed Dick A’huille, Nak’azdli First Nation Moise and Mary Antwoine, Saik’uz First Nation George George, Sr. Nadleh Whut’en First Nation Rita George, Wet’suwet’en First Nation Patrick Isaac, Wet’suwet’en First Nation Peter John, Burns Lake Band Alma Larson, Wet’suwet’en First Nation Betsy and Carl Leon, Nak’azdli First Nation Bernadette McQuarry, Nadleh Whut’en First Nation Aileen Prince, Nak’azdli First Nation Donald Prince, Nak’azdli First Nation Guy Prince, Nak’azdli First Nation Vince Prince, Nak’azdli First Nation Kenny Sam, Burns Lake Band Lillian Sam, Nak’azdli First Nation Ruth Tibbetts, Burns Lake Band Ryan Tibbetts, Burns Lake Band Joseph Tom, Wet’suwet’en First Nation Translation services provided by Lillian Morris, Wet’suwet’en First Nation. -

Food Web E.2. Electronic Appendix
E.2. Electronic Appendix - Food Web Elements of the Fraser River Basin Upper River (above rkm 210) Food webs: Microbenthic algae (periphyton), detritus from riparian vegetation and littoral insects (especially midges) are key components supporting fish production in the mainstem upper Fraser and larger tributaries. Collector-gatherers (invertebrates feeding on fine particulate organic material) are the most abundant functional feeding group, making up to 85% of the invertebrate species on the latter two rivers. Smaller tributaries are dominated by collector, shredder and grazer insect feeding modes (Reece and Richardson 2000). There is a general increasing trend in insect abundance from the headwaters of the main river to the lower river (Reynoldson et al. 2005). Juvenile stream-type Chinook rear along the shorelines of the upper river and tributaries and some overwinter under ice as the river margins usually freeze over here. Juvenile Chinook diets in the main stem and tributaries include larval plecopterans, empheropterans, chironomids and terrestrial insects (Homoptera, Coleoptera, Hymenoptera and Arachnida; Russell et al. 1983, Rogers et al. 1988, Levings and Lauzier 1991). Rainbow trout and northern pikeminnow consume mainly sculpins in the Nechako River as well as a variety of insects (Brown et al. 1992). Stressors: Water quality and habitat conditions have changed food webs in specific locations in the upper river. However, compared to other rivers in North America, water quality is good (Reynoldson et al. 2005), even with five pulp mills currently operating in the megareach. The food web of the Thompson River was stimulated in the past by low concentrations of bleached Kraft pulp mill effluent released into the river (Dube and Culp 1997); it is not known if this is still happening as treatment techniques for effluent have changed. -

Habitat-Based Methods to Estimate Spawner Capacity for Chinook
C S A S S C C S Canadian Science Advisory Secretariat Secrétariat canadien de consultation scientifique Research Document 2002/114 Document de recherche 2002/114 Not to be cited without Ne pas citer sans permission of the authors * autorisation des auteurs * Habitat-based methods to estimate Méthodes axées sur l’habitat pour estimer spawner capacity for chinook salmon in la capacité d’accueil de saumons the Fraser River watershed quinnats géniteurs dans le réseau fluvial du fleuve Fraser C. K. Parken1 , J. R. Irvine1, R. E. Bailey2, I. V. Williams3 1Fisheries and Oceans Canada Science Branch, Stock Assessment Division Pacific Biological Station Nanaimo, B.C. V9T 6N7 2Fisheries and Oceans Canada BC Interior, Resource Management 1278 Dalhousie Drive Kamloops, B.C. V2B 6G3 3I.V. Williams Consulting Ltd. 3565 Planta Rd. Nanaimo, B.C. V9T 1M1 * This series documents the scientific basis for the * La présente série documente les bases scientifiques evaluation of fisheries resources in Canada. As such, des évaluations des ressources halieutiques du Canada. it addresses the issues of the day in the time frames Elle traite des problèmes courants selon les échéanciers required and the documents it contains are not dictés. Les documents qu’elle contient ne doivent pas intended as definitive statements on the subjects être considérés comme des énoncés définitifs sur les addressed but rather as progress reports on ongoing sujets traités, mais plutôt comme des rapports d’étape investigations. sur les études en cours. Research documents are produced in the official Les documents de recherche sont publiés dans la language in which they are provided to the langue officielle utilisée dans le manuscrit envoyé au Secretariat. -
Fish 2002 Tec Doc Draft3
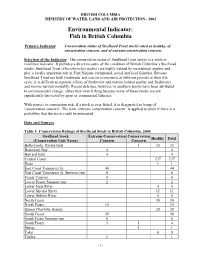
BRITISH COLUMBIA MINISTRY OF WATER, LAND AND AIR PROTECTION - 2002 Environmental Indicator: Fish in British Columbia Primary Indicator: Conservation status of Steelhead Trout stocks rated as healthy, of conservation concern, and of extreme conservation concern. Selection of the Indicator: The conservation status of Steelhead Trout stocks is a state or condition indicator. It provides a direct measure of the condition of British Columbia’s Steelhead stocks. Steelhead Trout (Oncorhynchus mykiss) are highly valued by recreational anglers and play a locally important role in First Nations ceremonial, social and food fisheries. Because Steelhead Trout use both freshwater and marine ecosystems at different periods in their life cycle, it is difficult to separate effects of freshwater and marine habitat quality and freshwater and marine harvest mortality. Recent delcines, however, in southern stocks have been attributed to environmental change, rather than over-fishing because many of these stocks are not significantly harvested by sport or commercial fisheries. With respect to conseration risk, if a stock is over fished, it is designated as being of ‘conservation concern’. The term ‘extreme conservation concern’ is applied to stock if there is a probablity that the stock could be extirpated. Data and Sources: Table 1. Conservation Ratings of Steelhead Stock in British Columbia, 2000 Steelhead Stock Extreme Conservation Conservation Healthy Total (Conservation Unit Name) Concern Concern Bella Coola–Rivers Inlet 1 32 33 Boundary Bay 4 4 Burrard -

Water Quality in British Columbia
WATER and AIR MONITORING and REPORTING SECTION WATER, AIR and CLIMATE CHANGE BRANCH MINISTRY OF ENVIRONMENT Water Quality in British Columbia _______________ Objectives Attainment in 2004 Prepared by: Burke Phippen BWP Consulting Inc. November 2005 WATER QUALITY IN B.C. – OBJECTIVES ATTAINMENT IN 2004 Canadian Cataloguing in Publication Data Main entry under title: Water quality in British Columbia : Objectives attainment in ... -- 2004 -- Annual. Continues: The Attainment of ambient water quality objectives. ISNN 1194-515X ISNN 1195-6550 = Water quality in British Columbia 1. Water quality - Standards - British Columbia - Periodicals. I. B.C. Environment. Water Management Branch. TD227.B7W37 363.73’942’0218711 C93-092392-8 ii WATER, AIR AND CLIMATE CHANGE BRANCH – MINISTRY OF ENVIRONMENT WATER QUALITY IN B.C. – OBJECTIVES ATTAINMENT IN 2004 TABLE OF CONTENTS TABLE OF CONTENTS......................................................................................................... III LIST OF TABLES .................................................................................................................. VI LIST OF FIGURES................................................................................................................ VII SUMMARY ........................................................................................................................... 1 ACKNOWLEDGEMENTS....................................................................................................... 2 INTRODUCTION.................................................................................................................. -

Lheidli T'enneh Perspectives on Resource Development
THE PARADOX OF DEVELOPMENT: LHEIDLI T'ENNEH PERSPECTIVES ON RESOURCE DEVELOPMENT by Geoffrey E.D. Hughes B.A., Northern Studies, University of Northern British Columbia, 2002 THESIS SUBMITTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF MASTER OF ARTS IN FIRST NATIONS STUDIES UNIVERSITY OF NORTHERN BRITISH COLUMBIA November 2011 © Geoffrey Hughes, 2011 Library and Archives Bibliotheque et Canada Archives Canada Published Heritage Direction du 1+1 Branch Patrimoine de I'edition 395 Wellington Street 395, rue Wellington Ottawa ON K1A0N4 Ottawa ON K1A 0N4 Canada Canada Your file Votre reference ISBN: 978-0-494-87547-6 Our file Notre reference ISBN: 978-0-494-87547-6 NOTICE: AVIS: The author has granted a non L'auteur a accorde une licence non exclusive exclusive license allowing Library and permettant a la Bibliotheque et Archives Archives Canada to reproduce, Canada de reproduire, publier, archiver, publish, archive, preserve, conserve, sauvegarder, conserver, transmettre au public communicate to the public by par telecommunication ou par I'lnternet, preter, telecommunication or on the Internet, distribuer et vendre des theses partout dans le loan, distrbute and sell theses monde, a des fins commerciales ou autres, sur worldwide, for commercial or non support microforme, papier, electronique et/ou commercial purposes, in microform, autres formats. paper, electronic and/or any other formats. The author retains copyright L'auteur conserve la propriete du droit d'auteur ownership and moral rights in this et des droits moraux qui protege cette these. Ni thesis. Neither the thesis nor la these ni des extraits substantiels de celle-ci substantial extracts from it may be ne doivent etre imprimes ou autrement printed or otherwise reproduced reproduits sans son autorisation. -

Nechako River Summary of Existing Data
Nechako Environmental Enhancement Fund British Columbia Nechako River Summary of Existing Data October 1999 Prepared by: RescanTM Environmental Services Ltd. Vancouver, British Columbia TM ERRATA Following are revisions to the Report “Nechako River – Summary of Existing Data” (October 1999) Page 2-13, Second Paragraph, Last Sentence: The last sentence in the second paragraph, i.e. “Design approval for the Cheslatta Fan Channel was obtained from the NFCP in July 1993”, should be replaced with the following: “The Nechako Fisheries Conservation Program gave a qualified approval for the design of a non-erodible channel (subject to nine items being addressed) and indicated final approval for the design of a channel would follow only after a regime channel concept had been considered further by the Kemano Completion Project and the Technical Committee.” Page 5-10, Figure 5-7: The top line in the Legend should read "Adult Sockeye Migration Routes". Page 5-15, Section 5.6.5, Waste Water Dilution: The two sentences at the bottom of this page should be replaced with the following: “Fort Fraser and Vanderhoof are the two main sources of sewage discharged to the Nechako River. Recently the firm, Urban Systems, was commissioned by Vanderhoof to undertake a study of their present sewage treatment system, which consists of retention lagoons, and to recommend options for enhancing treatment from this major point source in the future.” TABLE OF CONTENTS TM NECHAKO RIVER SUMMARY OF EXISTING DATA TABLE OF CONTENTS TABLE OF CONTENTS ................................................................................................................................i -

Sturgeon History
Sturgeon Calendar Sturgeon History UNEL’TSOO’S LIFE 1920s: Unel’tsoo, the oldest sturgeon that has been caught GEOGRAPHICAL TIME by the NWSRI, hatched when the Rightly called a pre-historic species, white sturgeon ancestors lived Nechako River flowed naturally. during the time of the dinosaurs, 170-245 million years ago! Physically, 1940s: Sturgeon were caught not much has changed. Their large, flat cartilaginous body covered and sold for food in stores in in sharp bony plates are features that developed a time long ago. Vanderhoof and Prince George. During the last ice age 50,000-10,000 years ago British Columbia was covered People also fished for sturgeon in ice a kilometre deep. When the ice receded it created new waterways all the way until the 1970s. across North America. Dakelh Elders tell the story of when the river flowed Unelt’soo eluded the anglers. backwards - when the Stuart River flowed north instead of south into the 1952: Kenney Dam was built on Nechako River as it does today. These kinds of ancient water connections the Nechako River. It changed and pathways allowed fish species to move great distances between the hydrology* of the river. continents and over mountains. The white sturgeon likely made their way to Unelt’soo survived the flow the Nechako River from the Upper Columbia River over 10,000 years ago. changes in the river. *study of the movement of water DISTRIBUTION > CLICK FOR THE NECHAKO WATERSHED MAP 1960s: Unelt’soo finally reached Globally, sturgeon are found circum-polar in the northern hemisphere. White maturity, however it is likely that sturgeon (Acipenser transmontanus) very few of Unel’tsoo’s eggs and live on the west coast of North larvae did not survive after 1967, America, and the Nechako white when researchers realized that sturgeon - a unique group within young fish were not surviving. -

Wild Rivers: Central British Columbia
Indian and Affaires indiennes Northern Affairs et du Nord Wild Rivers: Parks Canada Pares Canada Central British Columbia Published by Parks Canada under authority of the Hon. J. Hugh Faulkner, Minister of Indian and Northern Affairs, Ottawa, 1978 QS-7064-000-EE-A1 Les releves de la serie «Les rivieres sauvages» sont egalement publies en francais. Canada Canada metric metrique Metric Commission Canada has granted use of the National Symbol for Metric Conversion. Wild Rivers: Central British Columbia Wild Rivers Survey Parks Canada ARC Branch Planning Division Ottawa, 1978 2 Cariboo and Quesnel rivers: Ishpa Moun tain from Sandy Lake 3 'It is difficult to find in life any event and water, taken in the abstract, fail as which so effectually condenses intense completely to convey any idea of their nervous sensation into the shortest fierce embracings in the throes of a possible space of time as does the rapid as the fire burning quietly in a work of shooting, or running an im drawing-room fireplace fails to convey mense rapid. There is no toil, no heart the idea of a house wrapped and breaking labour about it, but as much sheeted in flames." coolness, dexterity, and skill as man can throw into the work of hand, eye Sir William Francis Butler (1872) and head; knowledge of when to strike and how to do it; knowledge of water and rock, and of the one hundred com binations which rock and water can assume — for these two things, rock 4 ©Minister of Supply and Services Now available in the Wild River Metric symbols used in this book Canada 1978 series: mm — millimetre(s) Available by mail from Printing and Alberta m — metre(s) Publishing, Supply and Services Central British Columbia km — kilometre(s) Canada, Ottawa, K1A 0S9, or through James Bay/Hudson Bay km/h - kilometres per hour your bookseller. -

Potential Effects of the Kekaho Completion Project on Fraser River Sockeye and Pink Salmon
POTENTIAL EFFECTS OF THE KEKAHO COMPLETION PROJECT ON FRASER RIVER SOCKEYE AND PINK SALMON by INTERNATIONAL PACIFIC SALMON FISHERIES COKHlSSION Appointed under a Convention Between Canada and the United States for the Protection, Preservation apd Extension of the Sockeye. and Pink Salmon Fisheries in the Fraser River Syste~ An analysis of potential effects of Alcan's 1983 expansion proposal on production of Fraser River sockeye and pink salmon is prelented 1n the above-noted 85-page report. This report, prepared by IPSFC, describes potential problems in the Nechako and Fraser Rivers that could .", affect sockeye and pink salmon production. A summary of the report is given in the following pages. Anyone requiring additional details may obtain a copy of the full report by writing to: International Pacific Salmon Fisheries Commission P. O. !lox 30 New Westminster, B.C. V3L 4X9 THE ALCAN PROPOSAL Alcan proposes to proceed with a second stage of development of tbe existing Nechako-Kemano hydro~ iectric diverlion. Initially, Alcan indicated the second stage would involve complete divers ion of the Necbako River above Kenney Dam but subsequently the Company has modified thia concept because of the various environmental concerna that were railed. The Company now proposes a schedule of flows to provide for other resource Ulera on Nechako River including ma intenance of habi t at for chinook salmon and tranaportation flow and r iver t eoperature control for protection of aockeye aallllOn. The proposed schedule i ncludes a continuous diacharge of 1100 cta lO Ve of 50°F water at the Nechako-Cheslatta confluence during the adult sockeye migration period and provision for flows up to 6000 cfs at 50°F, which would be uled in response to weather forecasts to limit the occurrencea of Nechako River t~eratures above Stuart River higher than 68°F and eli.inate occurrence of temperatures higher than 71°F. -

Prince George: Socio-Historical, Geographical, Political, and Economic Context Profile1
Prince George: Socio-historical, geographical, political, and economic context profile1 Prince George at night (Prince George Métis Community Association, 2014) Prepared by EQUIP Research staff on behalf of Central Interior Native Health Society Please Feel Free to Distribute Widely Colleen M. Varcoe, RN, PhD, and Annette J. Browne, RN, PhD Co-Principal Investigators Rochelle Einboden, MSN Research Assistant Acknowledgments: Thank you to Sarah de Leeuw, Carlene Dingwall, and Murry Krause for their input and review, to Kelsey Timler for her editorial and research support, and to Georgina Martin for her contributions to background materials. 1 Please cite this monograph as: Varcoe, C. M., Browne, A. J., & Einboden, R. (2015). Prince George: Socio- historical, geographical, political and economic context profile. EQUIP Healthcare: Research to equip primary healthcare for equity, in partnership with Central Interior Native Health Society. University of British Columbia, Vancouver & Prince George, BC. Version: February 19, 2015 This research program was supported by the Canadian Institutes of Health Research (www.cihr-irsc.gc.ca). Table of Contents Overview ..................................................................................................................................... 3 The Lheidli T’enneh Nation........................................................................................................ 4 The impacts of colonialism ........................................................................................................ -

Protecting Water Our Way
Protecting Water Our Way FIRST NATIONS FRESHWATER GOVERNANCE IN BRITISH COLUMBIA CONTENTS The First Nations Fisheries Council’s Water for Fish Freshwater Initiative ...............................................................1 Acknowledgements ................................................................................................................................................................ 2 About this Report ...................................................................................................................................................................4 Protecting Water Our Way: Introduction ..........................................................................................................................5 Whose Water is it, Anyway? A Word on Aboriginal Water Rights ...............................................................................8 First Nations-Led Freshwater Governance and Planning in British Columbia: Five Case Stories ........................ 14 1. Yinka Dene ‘Uza’hné Water Declaration and Policy Standards ........................................................................... 15 2. Syilx Nation and siw kw (Water) Declaration and Water Responsibility Planning Methodology ................ 19 3. Water Monitoring: Gateway to Governance ...........................................................................................................25 4. Tla’amin Nation and Negotiating Shared Decision Making in the Theodosia River Watershed .................29 5. Cowichan Tribes and the