The Components of Directional and Disruptive Selection in Heterogeneous Group-Structured Populations
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

And Ford, I; Ford, '953) on the Other Hand Have Put Forward a View Intermediate Between the Extreme Ones of Darwin on the One Hand and Goldschmidt on the Other
THE EVOLUTION OF MIMICRY IN THE BUTTERFLY PAPILIO DARDANUS C. A. CLARKE and P. M. SHEPPARD Departments of Medicine and Zoology, University of Liverpool Received23.V.59 1.INTRODUCTION WHENBatesputforward the mimicry hypothesis which bears his name, Darwin (1872), although accepting it, had some difficulty in explaining the evolution of the mimetic resemblance of several distinct species to one distasteful model by a series of small changes, a require- ment of his general theory of evolution. He said "it is necessary to suppose in some cases that ancient members belonging to several distinct groups, before they had diverged to their present extent, accidentally resembled a member of another and protected group in a sufficient degree to afford some slight protection; this having given the basis for the subsequent acquisition of the most perfect resemb- lance ". Punnett (1915) realised that the difficulty is even more acute when one is dealing with a polymorphic species whose forms mimic very distantly related models. Knowing that, in those butterflies which had been investigated genetically, the forms differed by single allelomorphs he concluded that the mimicry did not evolve gradually and did not confer any advantage or disadvantage to the individual. He argued that an allelomorph arises at a single step by mutation and that therefore the mimicry also arises by chance at a single step. Goldschmidt (x) although not denying that mimicry confers some advantage to its possessors also maintained that the resemblance arises fully perfected by a single mutation of a gene distinct from that producing the colour pattern in the model, but producing a similar effect in the mimic. -

Microevolution and the Genetics of Populations Microevolution Refers to Varieties Within a Given Type
Chapter 8: Evolution Lesson 8.3: Microevolution and the Genetics of Populations Microevolution refers to varieties within a given type. Change happens within a group, but the descendant is clearly of the same type as the ancestor. This might better be called variation, or adaptation, but the changes are "horizontal" in effect, not "vertical." Such changes might be accomplished by "natural selection," in which a trait within the present variety is selected as the best for a given set of conditions, or accomplished by "artificial selection," such as when dog breeders produce a new breed of dog. Lesson Objectives ● Distinguish what is microevolution and how it affects changes in populations. ● Define gene pool, and explain how to calculate allele frequencies. ● State the Hardy-Weinberg theorem ● Identify the five forces of evolution. Vocabulary ● adaptive radiation ● gene pool ● migration ● allele frequency ● genetic drift ● mutation ● artificial selection ● Hardy-Weinberg theorem ● natural selection ● directional selection ● macroevolution ● population genetics ● disruptive selection ● microevolution ● stabilizing selection ● gene flow Introduction Darwin knew that heritable variations are needed for evolution to occur. However, he knew nothing about Mendel’s laws of genetics. Mendel’s laws were rediscovered in the early 1900s. Only then could scientists fully understand the process of evolution. Microevolution is how individual traits within a population change over time. In order for a population to change, some things must be assumed to be true. In other words, there must be some sort of process happening that causes microevolution. The five ways alleles within a population change over time are natural selection, migration (gene flow), mating, mutations, or genetic drift. -

Patterns and Power of Phenotypic Selection in Nature
Articles Patterns and Power of Phenotypic Selection in Nature JOEL G. KINGSOLVER AND DAVID W. PFENNIG Phenotypic selection occurs when individuals with certain characteristics produce more surviving offspring than individuals with other characteristics. Although selection is regarded as the chief engine of evolutionary change, scientists have only recently begun to measure its action in the wild. These studies raise numerous questions: How strong is selection, and do different types of traits experience different patterns of selection? Is selection on traits that affect mating success as strong as selection on traits that affect survival? Does selection tend to favor larger body size, and, if so, what are its consequences? We explore these questions and discuss the pitfalls and future prospects of measuring selection in natural populations. Keywords: adaptive landscape, Cope’s rule, natural selection, rapid evolution, sexual selection henotypic selection occurs when individuals with selection on traits that affect survival stronger than on those Pdifferent characteristics (i.e., different phenotypes) that affect only mating success? In this article, we explore these differ in their survival, fecundity, or mating success. The idea and other questions about the patterns and power of phe- of phenotypic selection traces back to Darwin and Wallace notypic selection in nature. (1858), and selection is widely accepted as the primary cause of adaptive evolution within natural populations.Yet Darwin What is selection, and how does it work? never attempted to measure selection in nature, and in the Selection is the nonrandom differential survival or repro- century following the publication of On the Origin of Species duction of phenotypically different individuals. -

Arise by Chance As the Result of Mutation. They Therefore Suggest
THE EVOLUTION OF DOMINANCE UNDER DISRUPTIVE SELECTION C. A. CLARKE and P. Ni. SHEPPARD Department of Medicine and Department of Zoology, University of Liverpool Received6.iii.59 1.INTRODUCTION INa paper on the effects of disruptive selection, Mather (1955) pointed out that if there are two optimum values for a character and all others are less advantageous or disadvantageous there will be disruptive selection which can lead to the evolution of a polymorphism. Sheppard (1958) argued that where such selection is effective and the change from one optimum value to the other is switched by a single pair of allelomorphs there will be three genotypes but only two advantageous phenotypes. Consequently if dominance were absent initially it would be evolved as a result of the disruptive selection, the heterozygote and one of the homozygotes both coming to resemble one of the two optimum phenotypes (see Ford, 1955, on Tripharna comes). Thoday (1959) has shown by means of an artificial selection experiment that, even when a character is, at the beginning, controlled polygenically (sternopleural chaeta-number in Drosophila) and there is 50 per cent. gene exchange between the "high" and "low" selected sub-popu- lations, a polymorphism can evolve. The most fully understood examples of disruptive selection (other than sex) are provided by instances of Batesian Mimicry, where there are a number of distinct warningly coloured species, acting as models, which are mimicked by the polymorphic forms of a single more edible species. Fisher and Ford (see Ford, 1953) have argued that a suffi- ciently good resemblance between mimic and model is not likely to arise by chance as the result of mutation. -
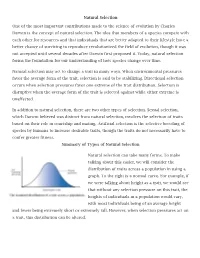
Natural Selection One of the Most Important Contributions Made to The
Natural Selection One of the most important contributions made to the science of evolution by Charles Darwin is the concept of natural selection. The idea that members of a species compete with each other for resources and that individuals that are better adapted to their lifestyle have a better chance of surviving to reproduce revolutionized the field of evolution, though it was not accepted until several decades after Darwin first proposed it. Today, natural selection forms the foundation for our understanding of how species change over time. Natural selection may act to change a trait in many ways. When environmental pressures favor the average form of the trait, selection is said to be stabilizing. Directional selection occurs when selection pressures favor one extreme of the trait distribution. Selection is disruptive when the average form of the trait is selected against while either extreme is unaffected. In addition to natural selection, there are two other types of selection. Sexual selection, which Darwin believed was distinct from natural selection, involves the selection of traits based on their role in courtship and mating. Artificial selection is the selective breeding of species by humans to increase desirable traits, though the traits do not necessarily have to confer greater fitness. Summary of Types of Natural Selection Natural selection can take many forms. To make talking about this easier, we will consider the distribution of traits across a population in using a graph. To the right is a normal curve. For example, if we were talking about height as a trait, we would see that without any selection pressure on this trait, the heights of individuals in a population would vary, with most individuals being of an average height and fewer being extremely short or extremely tall. -

Consequences of the Domestication of Man's Best Friend, The
Digital Comprehensive Summaries of Uppsala Dissertations from the Faculty of Science and Technology 289 Consequences of the Domestication of Man’s Best Friend, The Dog SUSANNE BJÖRNERFELDT ACTA UNIVERSITATIS UPSALIENSIS ISSN 1651-6214 UPPSALA ISBN 978-91-554-6854-5 2007 urn:nbn:se:uu:diva-7799 ! "# $ % &''( '!)'' * # * * +# #, "# - #, ./ * 0, &''(, 1 * # * $ 2 3 "# , 4 , &5!, 67 , , 809 !(5:!;:<<7:65<7:<, "# - # * # ;< ''' , " # # - # # = # , # - * # - * * # * * #, "# # * # # # # # 2 # # # # , " # - # - ) # # 94 # > # # , 8 1 # - # ** - # # # 2 * , 3 - * # - # # -# * , "# # * # # # * * * # # # , ? # * , * 1 * * - # *- * , @ - * # * - # -## * * , "# 1 # # # * * # * * , + 2 * # * - # - - # 2 , # > # -# * * - 1 * !" # $ # % # $ # & '( )*# # $+,-./0 # A 0 ./ * &''( 8009 ;6<;:6&;7 809 !(5:!;:<<7:65<7:< ) ))) :((!! B# )CC ,,C D E ) ))) :((!!F All knowledge, the totality of all questions and answers, is contained in the dog. FRANZ KAFKA, Investigations of the dog Cover photos by S Björnerfeldt List of Papers This thesis -

Biol2007 - Evolution of Quantitative Characters
1 BIOL2007 - EVOLUTION OF QUANTITATIVE CHARACTERS How do evolutionary biologists measure variation in a typical quantitative character? Let’s use beak size in birds as a typical example. Phenotypic variation (Vp) in a quantitative character Some of the variation in beak size is caused by environmental factors (VE). Some of the variation is caused by genetic factors (VG). If there was no variation in genotype between birds then individuals would differ in beak size only because of different rearing environments. Quantitative geneticists think in terms of how the value of a character in an individual compares to the mean value of the character in the population. The population mean is taken to be a neutral measure and then particular phenotypes are increases or decreases from that value. AT LEVEL OF INDIVIDUAL Phenotype = genotype + environment P = G + E AT LEVEL OF POPULATION Phenotypic variance = genotypic variance + environmental variance VP = VG + VE Genotypic variance is divided into additive, dominance and interaction components VG = VA + VD + V I VA = Additive variance – due to the additive effects of individual alleles (inherited) VD = Dominance variance - reflect the combinations of alleles at each locus (homo- versus (homozygotes versus hetero-zygotes) in any given generation but are not inherited by offspring. An intra-locus effect. VI = Interaction/epistatic variance - reflect epistatic interactions between alleles at different loci. Again not passed onto offspring; they depend on particular combinations of genes in a given generation and are not inherited by offspring. An inter-locus effect. ___________________________________________________________ The following section (italicised) provides the alebgra to backup my assertion that VD and VI are not heritable. -

FULLTEXT01.Pdf
Journal of Theoretical Biology 507 (2020) 110449 Contents lists available at ScienceDirect Journal of Theoretical Biology journal homepage: www.elsevier.com/locate/yjtbi The components of directional and disruptive selection in heterogeneous group-structured populations ⇑ Hisashi Ohtsuki a, , Claus Rueffler b, Joe Yuichiro Wakano c,d,e, Kalle Parvinen f,g, Laurent Lehmann h a Department of Evolutionary Studies of Biosystems, School of Advanced Sciences, SOKENDAI (The Graduate University for Advanced Studies), Shonan Village, Hayama, Kanagawa 240-0193, Japan b Animal Ecology, Department of Ecology and Genetics, University of Uppsala, Sweden c School of Interdisciplinary Mathematical Sciences, Meiji University, Tokyo 164-8525, Japan d Meiji Institute for Advanced Study of Mathematical Sciences, Tokyo 164-8525, Japan e Department of Mathematics, The University of British Columbia, 1984 Mathematics Road, Vancouver, B.C. V6T 1Z2, Canada f Department of Mathematics and Statistics, FI-20014 University of Turku, Finland g Evolution and Ecology Program, International Institute for Applied Systems Analysis (IIASA), A-2361 Laxenburg, Austria h Department of Ecology and Evolution, University of Lausanne, Switzerland article info abstract Article history: We derive how directional and disruptive selection operate on scalar traits in a heterogeneous group- Received 31 March 2020 structured population for a general class of models. In particular, we assume that each group in the Revised 5 August 2020 population can be in one of a finite number of states, where states can affect group size and/or other envi- Accepted 7 August 2020 ronmental variables, at a given time. Using up to second-order perturbation expansions of the invasion Available online 16 August 2020 fitness of a mutant allele, we derive expressions for the directional and disruptive selection coefficients, which are sufficient to classify the singular strategies of adaptive dynamics. -

What Is Natural Selection?
This week (June 8th – 12th) we are reviewing evolution. 1. Review the lecture on evolution 2. Watch the evidence of evolution video: https://www.khanacademy.org/science/high-school-biology/hs-evolution/hs- evidence-of-evolution/v/evidence-for-evolution 3. Watch the Types of Natural Selection video https://youtu.be/64JUJdZdDQo 4. Complete the Evolution Review Activity 5. Complete the Natural Selection Reading and Questions I am grading you based on the rubric below. If you get below a 70% I will allow you to redo the assignment. Evidence of Evolution & the Week 5 Remote Learning Mechanisms of Biodiversity Natural Selection The total variety of all the organisms in the biosphere= Biodiversity Where did all these different organisms come from? How are they related? Some organisms in a population are less likely to survive. Over time, natural selection results in changes in the inherited characteristics of a population. WHAT IS DARWIN’S THEORY? These changes increase a species’ fitness in its environment. Any inherited characteristic Ability of an individual to that increases an organism’s survive and reproduce in its chance of survival= specific environment= FITNESS ADAPTATION WHAT IS DARWIN’S THEORY? STRUGGLE FOR EXISTENCE: means that members of each species must compete for food, space, and other resources. SURVIVAL OF THE FITTEST: organisms which are better adapted to the environment will survive and reproduce, passing on their genes. DESCENT WITH MODIFICATION: suggests that each species has decended, with changes, from other species over time. • This idea suggests that all living species are related to each other, and that all species, living and extinct,share a common ancestor. -

Disruptive Selection and Then What?
Review TRENDS in Ecology and Evolution Vol.21 No.5 May 2006 Disruptive selection and then what? Claus Rueffler1,2, Tom J.M. Van Dooren2, Olof Leimar3 and Peter A. Abrams1 1Department of Zoology, University of Toronto, 25 Harbord Street, Toronto, Ontario, Canada, M5S 3G5 2Institute of Biology Leiden, Leiden University, Kaiserstraat 63, 2311 GP Leiden, the Netherlands 3Department of Zoology, Stockholm University, SE-106 91, Stockholm, Sweden Disruptive selection occurs when extreme phenotypes more extreme phenotypes. Thus, directional selection acts have a fitness advantage over more intermediate towards specialization in the direction of the closer phenotypes. The phenomenon is particularly interesting fitness peak. when selection keeps a population in a disruptive For the second type of disruptive selection, we can regime. This can lead to increased phenotypic variation imagine a situation where a population exploits a while disruptive selection itself is diminished or elimi- continuously varying resource, such as seeds that nated. Here, we review processes that increase pheno- range from very small to very large and where the typic variation in response to disruptive selection and level of consumption influences seed abundance. Indi- discuss some of the possible outcomes, such as viduals that efficiently exploit the most abundant sympatric species pairs, sexual dimorphisms, pheno- resource (e.g. seeds of intermediate size) have a fitness typic plasticity and altered community assemblages. We also identify factors influencing the likelihoods of these different outcomes. Glossary Assortative mating: when sexually reproducing organisms tend to mate with individuals that are similar to themselves in some respect. Can be caused by Introduction assortative mate choice, or by environmental factors that cause non-random A population experiences disruptive selection (see associations between mating partners. -

POPULATION GENETICS LECTURE NOTES-2016.Pdf
Training course in Quantitative Genetics and Genomics Biosciences East and Central Africa- International Livestock Research Institute (BecA-ILRI) Hub Nairobi, KENYA May 30-June 10, 2016 POPULATION AND QUANTITATIVE GENETICS GENOME ORGANIZATION AND GENETIC MARKERS SELECTION THEORY BREEDING STRATEGIES Samuel E Aggrey, PhD Professor Department of Poultry Science Institute of Bioinformatics University of Georgia Athens, GA 30602, USA [email protected] Preface This lecture notes was written in an attempt to cover parts of Population Genetics, Quantitative Genetics and Molecular Genetics for postgraduate students and also as a refresher for field geneticists. The course material is not a text book and not meant to be copied, duplicated or sold. This text is unedited and I am solely responsible for all conceptual mistakes, grammatical errors and typos. Genetics is a life-long course and cannot be covered in a few lectures. Only selected parts of the population- and quantitative-, and molecular genetics will be covered in this course because of time constraints. This course will cover some of the evolutionary changes in allele frequency between generations such as natural selection and gene flow, and some aspects of Quantitative and Molecular Genetics. To those men who have kept us awake for over two centuries and I believe would continue to do so for many more centuries! POPULATION GENETICS The study of composition of biological populations, and changes in genetic composition that result from operation of various factors including (a) natural selection, (b) genetic drift, (c) mutations and (d) gene flow Genetic composition Population 1. The number of alleles at a locus A group of breeding 2. -

Population Genetics Simulations Heath Blackmon and Emma E
Population Genetics Simulations Heath Blackmon and Emma E. Goldberg last updated: 2016-04-02 Contents Introduction 1 Evolution Across Generations.......................................1 Lauching the Population Genetics Application.............................2 Selection 3 Directional selection............................................3 Overdominance...............................................4 Underdominance..............................................4 Genetic Drift 5 Drift Alone.................................................5 Drift and Selection.............................................5 Mutation 6 Introduction Once an allele enters a population (by mutation or migration), its evolutionary fate is determined by selection and drift. In this lab, you will use a computer program to explore how the the prevalence of an allele in a population (its frequency) is affected by a variety of factors—its initial frequency, whether it is dominant or recessive, the relative fitnesses of individuals bearing different alleles, and population size. Throughout this lab, we will focus on a single locus with two alleles, A and a. We will assume that mating is random with respect to this locus, and that there is no migration from other population. Evolution Across Generations The core of a population genetics model is to describe how allele frequencies change over time. A computer can quickly simulate many generations, rather than doing a ton of calculations by hand. We will use an R computer program in this lab. You don’t need to worry about the actual code that it contains,however, it is important to understand what goes on inside such a program. Let’s focus on the scenario in which there is selection, but not drift, mutation, or migration. The basic idea is to start with some initial frequency of allele A, compute how it changes within the first generation, and subsequent generations.