Population Genetics Simulations Heath Blackmon and Emma E
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

And Ford, I; Ford, '953) on the Other Hand Have Put Forward a View Intermediate Between the Extreme Ones of Darwin on the One Hand and Goldschmidt on the Other
THE EVOLUTION OF MIMICRY IN THE BUTTERFLY PAPILIO DARDANUS C. A. CLARKE and P. M. SHEPPARD Departments of Medicine and Zoology, University of Liverpool Received23.V.59 1.INTRODUCTION WHENBatesputforward the mimicry hypothesis which bears his name, Darwin (1872), although accepting it, had some difficulty in explaining the evolution of the mimetic resemblance of several distinct species to one distasteful model by a series of small changes, a require- ment of his general theory of evolution. He said "it is necessary to suppose in some cases that ancient members belonging to several distinct groups, before they had diverged to their present extent, accidentally resembled a member of another and protected group in a sufficient degree to afford some slight protection; this having given the basis for the subsequent acquisition of the most perfect resemb- lance ". Punnett (1915) realised that the difficulty is even more acute when one is dealing with a polymorphic species whose forms mimic very distantly related models. Knowing that, in those butterflies which had been investigated genetically, the forms differed by single allelomorphs he concluded that the mimicry did not evolve gradually and did not confer any advantage or disadvantage to the individual. He argued that an allelomorph arises at a single step by mutation and that therefore the mimicry also arises by chance at a single step. Goldschmidt (x) although not denying that mimicry confers some advantage to its possessors also maintained that the resemblance arises fully perfected by a single mutation of a gene distinct from that producing the colour pattern in the model, but producing a similar effect in the mimic. -

Microevolution and the Genetics of Populations Microevolution Refers to Varieties Within a Given Type
Chapter 8: Evolution Lesson 8.3: Microevolution and the Genetics of Populations Microevolution refers to varieties within a given type. Change happens within a group, but the descendant is clearly of the same type as the ancestor. This might better be called variation, or adaptation, but the changes are "horizontal" in effect, not "vertical." Such changes might be accomplished by "natural selection," in which a trait within the present variety is selected as the best for a given set of conditions, or accomplished by "artificial selection," such as when dog breeders produce a new breed of dog. Lesson Objectives ● Distinguish what is microevolution and how it affects changes in populations. ● Define gene pool, and explain how to calculate allele frequencies. ● State the Hardy-Weinberg theorem ● Identify the five forces of evolution. Vocabulary ● adaptive radiation ● gene pool ● migration ● allele frequency ● genetic drift ● mutation ● artificial selection ● Hardy-Weinberg theorem ● natural selection ● directional selection ● macroevolution ● population genetics ● disruptive selection ● microevolution ● stabilizing selection ● gene flow Introduction Darwin knew that heritable variations are needed for evolution to occur. However, he knew nothing about Mendel’s laws of genetics. Mendel’s laws were rediscovered in the early 1900s. Only then could scientists fully understand the process of evolution. Microevolution is how individual traits within a population change over time. In order for a population to change, some things must be assumed to be true. In other words, there must be some sort of process happening that causes microevolution. The five ways alleles within a population change over time are natural selection, migration (gene flow), mating, mutations, or genetic drift. -

Polymorphism Under Apostatic
Heredity (1984), 53(3), 677—686 1984. The Genetical Society of Great Britain POLYMORPHISMUNDER APOSTATIC AND APOSEMATIC SELECTION VINTON THOMPSON Department of Biology, Roosevelt University, 430 South Michigan Avenue, Chicago, Illinois 60805 USA Received31 .i.84 SUMMARY Selection for warning colouration in well-defended species should lead to a single colour form in each local population, but some species are locally polymor- phic for aposematic colour forms. Single-locus two-allele models of frequency- dependent selection indicate that combined apostatic and aposematic selection may maintain stable polymorphism for one, two or three aposematic forms, provided that at least one form is subject to net apostatic selection. Frequency- independent selective differences between colour forms broaden the possibilities for aposematic polymorphism but lead to monomorphism if too large. Concurrent apostatic and aposematic selection may explain polymorphism for warning colouration in a number of jumping or moderately unpalatable insects. 1. INTRODUCTION Polymorphism for warning colouration poses a paradox for evolutionists because, as Fisher (1958) seems to have been the first to note, selection for warning colouration (aposematic selection) should lead to monomorphism. Under aposematic selection predators tend to avoid previously encountered phenotypes of undesirable prey, so that the fitness of each phenotype increases as it becomes more common. Given the presence of two forms, each will suffer at the hands of predators conditioned to avoid the other and one will eventually prevail because as soon as it gets the upper hand numerically it will drive the other to extinction. Nonetheless, many species are polymorphic for colour forms that appear to be aposematic. The ladybird beetles (Coccinellidae) furnish a number of particularly striking examples (Hodek, 1973; and see illustration in Ayala, 1978). -

Patterns and Power of Phenotypic Selection in Nature
Articles Patterns and Power of Phenotypic Selection in Nature JOEL G. KINGSOLVER AND DAVID W. PFENNIG Phenotypic selection occurs when individuals with certain characteristics produce more surviving offspring than individuals with other characteristics. Although selection is regarded as the chief engine of evolutionary change, scientists have only recently begun to measure its action in the wild. These studies raise numerous questions: How strong is selection, and do different types of traits experience different patterns of selection? Is selection on traits that affect mating success as strong as selection on traits that affect survival? Does selection tend to favor larger body size, and, if so, what are its consequences? We explore these questions and discuss the pitfalls and future prospects of measuring selection in natural populations. Keywords: adaptive landscape, Cope’s rule, natural selection, rapid evolution, sexual selection henotypic selection occurs when individuals with selection on traits that affect survival stronger than on those Pdifferent characteristics (i.e., different phenotypes) that affect only mating success? In this article, we explore these differ in their survival, fecundity, or mating success. The idea and other questions about the patterns and power of phe- of phenotypic selection traces back to Darwin and Wallace notypic selection in nature. (1858), and selection is widely accepted as the primary cause of adaptive evolution within natural populations.Yet Darwin What is selection, and how does it work? never attempted to measure selection in nature, and in the Selection is the nonrandom differential survival or repro- century following the publication of On the Origin of Species duction of phenotypically different individuals. -

Signals of Predation-Induced Directional and Disruptive Selection in the Threespine Stickleback
Eawag_07048 Evolutionary Ecology Research, 2012, 14: 193–205 Signals of predation-induced directional and disruptive selection in the threespine stickleback Michael Zeller, Kay Lucek, Marcel P. Haesler, Ole Seehausen and Arjun Sivasundar Institute for Ecology and Evolution, University of Bern, Bern, Switzerland and Department of Fish Ecology, Eawag Centre for Ecology, Evolution and Biogeochemistry, Kastanienbaum, Switzerland ABSTRACT Background: Different predation regimes may exert divergent selection pressure on phenotypes and their associated genotypes. Threespine stickleback Gasterosteus aculeatus have a suite of bony structures, which have been shown to be an effective defence against predation and have a well-known genetic basis. Question: Do different predator regimes induce different selective pressures on growth rates and defence phenotypes in threespine stickleback between different habitats across distinct age classes? Hypothesis: In the presence of predation-induced selection, we expect diverging morphological responses between populations experiencing either low or high predation pressure. Study system: Threespine stickleback were sampled from two natural but recently established populations in an invasive range. One site has a high density of fish and insect predators, while at the other site predation pressure is low. Methods: We inferred predator-induced selection on defence traits by comparing the distribution of size classes, defence phenotypes, and an armour-related genotype between dif- ferent age classes in a high and a low predation regime. Results: Under high predation, there are indications of directional selection for faster growth, whereas lateral plate phenotypes and associated genotypes show indications for disruptive selection. Heterozygotes at the Eda-gene have a lower survival rate than either homozygote. Neither pattern is evident in the low predation regime. -

Arise by Chance As the Result of Mutation. They Therefore Suggest
THE EVOLUTION OF DOMINANCE UNDER DISRUPTIVE SELECTION C. A. CLARKE and P. Ni. SHEPPARD Department of Medicine and Department of Zoology, University of Liverpool Received6.iii.59 1.INTRODUCTION INa paper on the effects of disruptive selection, Mather (1955) pointed out that if there are two optimum values for a character and all others are less advantageous or disadvantageous there will be disruptive selection which can lead to the evolution of a polymorphism. Sheppard (1958) argued that where such selection is effective and the change from one optimum value to the other is switched by a single pair of allelomorphs there will be three genotypes but only two advantageous phenotypes. Consequently if dominance were absent initially it would be evolved as a result of the disruptive selection, the heterozygote and one of the homozygotes both coming to resemble one of the two optimum phenotypes (see Ford, 1955, on Tripharna comes). Thoday (1959) has shown by means of an artificial selection experiment that, even when a character is, at the beginning, controlled polygenically (sternopleural chaeta-number in Drosophila) and there is 50 per cent. gene exchange between the "high" and "low" selected sub-popu- lations, a polymorphism can evolve. The most fully understood examples of disruptive selection (other than sex) are provided by instances of Batesian Mimicry, where there are a number of distinct warningly coloured species, acting as models, which are mimicked by the polymorphic forms of a single more edible species. Fisher and Ford (see Ford, 1953) have argued that a suffi- ciently good resemblance between mimic and model is not likely to arise by chance as the result of mutation. -
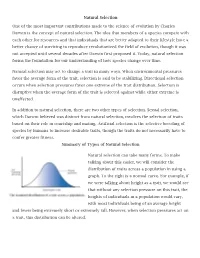
Natural Selection One of the Most Important Contributions Made to The
Natural Selection One of the most important contributions made to the science of evolution by Charles Darwin is the concept of natural selection. The idea that members of a species compete with each other for resources and that individuals that are better adapted to their lifestyle have a better chance of surviving to reproduce revolutionized the field of evolution, though it was not accepted until several decades after Darwin first proposed it. Today, natural selection forms the foundation for our understanding of how species change over time. Natural selection may act to change a trait in many ways. When environmental pressures favor the average form of the trait, selection is said to be stabilizing. Directional selection occurs when selection pressures favor one extreme of the trait distribution. Selection is disruptive when the average form of the trait is selected against while either extreme is unaffected. In addition to natural selection, there are two other types of selection. Sexual selection, which Darwin believed was distinct from natural selection, involves the selection of traits based on their role in courtship and mating. Artificial selection is the selective breeding of species by humans to increase desirable traits, though the traits do not necessarily have to confer greater fitness. Summary of Types of Natural Selection Natural selection can take many forms. To make talking about this easier, we will consider the distribution of traits across a population in using a graph. To the right is a normal curve. For example, if we were talking about height as a trait, we would see that without any selection pressure on this trait, the heights of individuals in a population would vary, with most individuals being of an average height and fewer being extremely short or extremely tall. -

Consequences of the Domestication of Man's Best Friend, The
Digital Comprehensive Summaries of Uppsala Dissertations from the Faculty of Science and Technology 289 Consequences of the Domestication of Man’s Best Friend, The Dog SUSANNE BJÖRNERFELDT ACTA UNIVERSITATIS UPSALIENSIS ISSN 1651-6214 UPPSALA ISBN 978-91-554-6854-5 2007 urn:nbn:se:uu:diva-7799 ! "# $ % &''( '!)'' * # * * +# #, "# - #, ./ * 0, &''(, 1 * # * $ 2 3 "# , 4 , &5!, 67 , , 809 !(5:!;:<<7:65<7:<, "# - # * # ;< ''' , " # # - # # = # , # - * # - * * # * * #, "# # * # # # # # 2 # # # # , " # - # - ) # # 94 # > # # , 8 1 # - # ** - # # # 2 * , 3 - * # - # # -# * , "# # * # # # * * * # # # , ? # * , * 1 * * - # *- * , @ - * # * - # -## * * , "# 1 # # # * * # * * , + 2 * # * - # - - # 2 , # > # -# * * - 1 * !" # $ # % # $ # & '( )*# # $+,-./0 # A 0 ./ * &''( 8009 ;6<;:6&;7 809 !(5:!;:<<7:65<7:< ) ))) :((!! B# )CC ,,C D E ) ))) :((!!F All knowledge, the totality of all questions and answers, is contained in the dog. FRANZ KAFKA, Investigations of the dog Cover photos by S Björnerfeldt List of Papers This thesis -

Population Dynamics of Underdominance Gene Drive Systems in Continuous Space
bioRxiv preprint doi: https://doi.org/10.1101/449355; this version posted October 22, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Population dynamics of underdominance gene drive systems in continuous space Jackson Champer1,2*, Joanna Zhao1, Samuel E. Champer1, Jingxian Liu1, Philipp W. Messer1* 1Department of Biological Statistics anD Computational Biology, 2Department of Molecular Biology anD Genetics, Cornell University, Ithaca, NY 14853 *CorresponDing authors: JC ([email protected]), PWM ([email protected]) ABSTRACT UnDerDominance gene Drive systems promise a mechanism for rapiDly spreaDing payloaD alleles through a local population while otherwise remaining confineD, unable to spreaD into neighboring populations due to their frequency-dependent Dynamics. Such systems coulD proviDe a new tool in the fight against vector-borne diseases by disseminating transgenic payloads through vector populations. If local confinement can inDeeD be achieveD, the decision-making process for the release of such constructs woulD likely be consiDerably simpler compareD to other gene Drive mechanisms such as CRISPR homing drives. So far, the confinement ability of underdominance systems has only been demonstrated in moDels of panmictic populations linkeD by migration. How such systems woulD behave in realistic populations where individuals move over continuous space remains largely unknown. Here, we stuDy several underdominance systems in continuous-space population models and show that their dynamics are drastically altered from those in panmictic populations. Specifically, we find that all underdominance systems we stuDied can fail to persist in such environments, even after successful local establishment. -

Biol2007 - Evolution of Quantitative Characters
1 BIOL2007 - EVOLUTION OF QUANTITATIVE CHARACTERS How do evolutionary biologists measure variation in a typical quantitative character? Let’s use beak size in birds as a typical example. Phenotypic variation (Vp) in a quantitative character Some of the variation in beak size is caused by environmental factors (VE). Some of the variation is caused by genetic factors (VG). If there was no variation in genotype between birds then individuals would differ in beak size only because of different rearing environments. Quantitative geneticists think in terms of how the value of a character in an individual compares to the mean value of the character in the population. The population mean is taken to be a neutral measure and then particular phenotypes are increases or decreases from that value. AT LEVEL OF INDIVIDUAL Phenotype = genotype + environment P = G + E AT LEVEL OF POPULATION Phenotypic variance = genotypic variance + environmental variance VP = VG + VE Genotypic variance is divided into additive, dominance and interaction components VG = VA + VD + V I VA = Additive variance – due to the additive effects of individual alleles (inherited) VD = Dominance variance - reflect the combinations of alleles at each locus (homo- versus (homozygotes versus hetero-zygotes) in any given generation but are not inherited by offspring. An intra-locus effect. VI = Interaction/epistatic variance - reflect epistatic interactions between alleles at different loci. Again not passed onto offspring; they depend on particular combinations of genes in a given generation and are not inherited by offspring. An inter-locus effect. ___________________________________________________________ The following section (italicised) provides the alebgra to backup my assertion that VD and VI are not heritable. -

FULLTEXT01.Pdf
Journal of Theoretical Biology 507 (2020) 110449 Contents lists available at ScienceDirect Journal of Theoretical Biology journal homepage: www.elsevier.com/locate/yjtbi The components of directional and disruptive selection in heterogeneous group-structured populations ⇑ Hisashi Ohtsuki a, , Claus Rueffler b, Joe Yuichiro Wakano c,d,e, Kalle Parvinen f,g, Laurent Lehmann h a Department of Evolutionary Studies of Biosystems, School of Advanced Sciences, SOKENDAI (The Graduate University for Advanced Studies), Shonan Village, Hayama, Kanagawa 240-0193, Japan b Animal Ecology, Department of Ecology and Genetics, University of Uppsala, Sweden c School of Interdisciplinary Mathematical Sciences, Meiji University, Tokyo 164-8525, Japan d Meiji Institute for Advanced Study of Mathematical Sciences, Tokyo 164-8525, Japan e Department of Mathematics, The University of British Columbia, 1984 Mathematics Road, Vancouver, B.C. V6T 1Z2, Canada f Department of Mathematics and Statistics, FI-20014 University of Turku, Finland g Evolution and Ecology Program, International Institute for Applied Systems Analysis (IIASA), A-2361 Laxenburg, Austria h Department of Ecology and Evolution, University of Lausanne, Switzerland article info abstract Article history: We derive how directional and disruptive selection operate on scalar traits in a heterogeneous group- Received 31 March 2020 structured population for a general class of models. In particular, we assume that each group in the Revised 5 August 2020 population can be in one of a finite number of states, where states can affect group size and/or other envi- Accepted 7 August 2020 ronmental variables, at a given time. Using up to second-order perturbation expansions of the invasion Available online 16 August 2020 fitness of a mutant allele, we derive expressions for the directional and disruptive selection coefficients, which are sufficient to classify the singular strategies of adaptive dynamics. -

Simulating Natural Selection in Landscape Genetics
Molecular Ecology Resources (2012) 12, 363–368 doi: 10.1111/j.1755-0998.2011.03075.x Simulating natural selection in landscape genetics E. L. LANDGUTH,* S. A. CUSHMAN† and N. A. JOHNSON‡ *Division of Biological Sciences, University of Montana, Missoula, MT 59812, USA, †Rocky Mountain Research Station, United States Forest Service, Flagstaff, AZ 86001, USA, USA, ‡Department of Plant, Soil, and Insect Sciences and Graduate Program in Organismic and Evolutionary Biology, University of Massachusetts, Amherst, MA 01003, USA Abstract Linking landscape effects to key evolutionary processes through individual organism movement and natural selection is essential to provide a foundation for evolutionary landscape genetics. Of particular importance is determining how spa- tially-explicit, individual-based models differ from classic population genetics and evolutionary ecology models based on ideal panmictic populations in an allopatric setting in their predictions of population structure and frequency of fixation of adaptive alleles. We explore initial applications of a spatially-explicit, individual-based evolutionary landscape genetics program that incorporates all factors – mutation, gene flow, genetic drift and selection – that affect the frequency of an allele in a population. We incorporate natural selection by imposing differential survival rates defined by local relative fitness values on a landscape. Selection coefficients thus can vary not only for genotypes, but also in space as functions of local environmental variability. This simulator enables coupling of gene flow (governed by resistance surfaces), with natural selection (governed by selection surfaces). We validate the individual-based simulations under Wright-Fisher assumptions. We show that under isolation-by-distance processes, there are deviations in the rate of change and equilibrium values of allele frequency.