Local Adaptation When Competition Depends on Phenotypic Similarity
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

And Ford, I; Ford, '953) on the Other Hand Have Put Forward a View Intermediate Between the Extreme Ones of Darwin on the One Hand and Goldschmidt on the Other
THE EVOLUTION OF MIMICRY IN THE BUTTERFLY PAPILIO DARDANUS C. A. CLARKE and P. M. SHEPPARD Departments of Medicine and Zoology, University of Liverpool Received23.V.59 1.INTRODUCTION WHENBatesputforward the mimicry hypothesis which bears his name, Darwin (1872), although accepting it, had some difficulty in explaining the evolution of the mimetic resemblance of several distinct species to one distasteful model by a series of small changes, a require- ment of his general theory of evolution. He said "it is necessary to suppose in some cases that ancient members belonging to several distinct groups, before they had diverged to their present extent, accidentally resembled a member of another and protected group in a sufficient degree to afford some slight protection; this having given the basis for the subsequent acquisition of the most perfect resemb- lance ". Punnett (1915) realised that the difficulty is even more acute when one is dealing with a polymorphic species whose forms mimic very distantly related models. Knowing that, in those butterflies which had been investigated genetically, the forms differed by single allelomorphs he concluded that the mimicry did not evolve gradually and did not confer any advantage or disadvantage to the individual. He argued that an allelomorph arises at a single step by mutation and that therefore the mimicry also arises by chance at a single step. Goldschmidt (x) although not denying that mimicry confers some advantage to its possessors also maintained that the resemblance arises fully perfected by a single mutation of a gene distinct from that producing the colour pattern in the model, but producing a similar effect in the mimic. -

Microevolution and the Genetics of Populations Microevolution Refers to Varieties Within a Given Type
Chapter 8: Evolution Lesson 8.3: Microevolution and the Genetics of Populations Microevolution refers to varieties within a given type. Change happens within a group, but the descendant is clearly of the same type as the ancestor. This might better be called variation, or adaptation, but the changes are "horizontal" in effect, not "vertical." Such changes might be accomplished by "natural selection," in which a trait within the present variety is selected as the best for a given set of conditions, or accomplished by "artificial selection," such as when dog breeders produce a new breed of dog. Lesson Objectives ● Distinguish what is microevolution and how it affects changes in populations. ● Define gene pool, and explain how to calculate allele frequencies. ● State the Hardy-Weinberg theorem ● Identify the five forces of evolution. Vocabulary ● adaptive radiation ● gene pool ● migration ● allele frequency ● genetic drift ● mutation ● artificial selection ● Hardy-Weinberg theorem ● natural selection ● directional selection ● macroevolution ● population genetics ● disruptive selection ● microevolution ● stabilizing selection ● gene flow Introduction Darwin knew that heritable variations are needed for evolution to occur. However, he knew nothing about Mendel’s laws of genetics. Mendel’s laws were rediscovered in the early 1900s. Only then could scientists fully understand the process of evolution. Microevolution is how individual traits within a population change over time. In order for a population to change, some things must be assumed to be true. In other words, there must be some sort of process happening that causes microevolution. The five ways alleles within a population change over time are natural selection, migration (gene flow), mating, mutations, or genetic drift. -

Gene Flow by Pollen Into Small Populations
Proc. Natl. Acad. Sci. USA Vol. 86, pp. 9044-9047, November 1989 Population Biology Gene flow by pollen into small populations: Data from experimental and natural stands of wild radish (Raphanus sativus/spatial isolation/interpopulation mating) NORMAN C. ELLSTRANDt, B. DEVLINt, AND DIANE L. MARSHALL§ Department of Botany and Plant Sciences and Program in Genetics, University of California, Riverside, CA 92521-0124 Communicated by R. W. Allard, August 14, 1989 (received for review August 18, 1988) ABSTRACT Gene flow can have an especially strong im- much from population to population, whether it varies within pact on the evolution of small populations. However, empirical a population over a season, and whether it varies with spatial studies on the actual rates and patterns of gene flow into small isolation from the nearest conspecific. More data are needed populations are few. Thus, we sought to measure gene flow into that focus more closely on a number ofpopulations ofa single small populations of wild radish, Raphanus sativus. We found species. significant differences in gene flow receipt among experimental Therefore, we selected wild radish, Raphanus sativus L. populations and within those populations over a season. A (Brassicaceae), to measure patterns of gene flow by pollen maximum-likelihood estimate revealed that almost all of the into small populations. This species is especially suitable for gene flow into these synthetic populations had its origin in both experimental and descriptive gene flow studies because relatively distant (>650 m), large natural populations rather it is a common outcrossing weed in southern California (8), than the proximal (255400 m), small synthetic populations. -

Polymorphism Under Apostatic
Heredity (1984), 53(3), 677—686 1984. The Genetical Society of Great Britain POLYMORPHISMUNDER APOSTATIC AND APOSEMATIC SELECTION VINTON THOMPSON Department of Biology, Roosevelt University, 430 South Michigan Avenue, Chicago, Illinois 60805 USA Received31 .i.84 SUMMARY Selection for warning colouration in well-defended species should lead to a single colour form in each local population, but some species are locally polymor- phic for aposematic colour forms. Single-locus two-allele models of frequency- dependent selection indicate that combined apostatic and aposematic selection may maintain stable polymorphism for one, two or three aposematic forms, provided that at least one form is subject to net apostatic selection. Frequency- independent selective differences between colour forms broaden the possibilities for aposematic polymorphism but lead to monomorphism if too large. Concurrent apostatic and aposematic selection may explain polymorphism for warning colouration in a number of jumping or moderately unpalatable insects. 1. INTRODUCTION Polymorphism for warning colouration poses a paradox for evolutionists because, as Fisher (1958) seems to have been the first to note, selection for warning colouration (aposematic selection) should lead to monomorphism. Under aposematic selection predators tend to avoid previously encountered phenotypes of undesirable prey, so that the fitness of each phenotype increases as it becomes more common. Given the presence of two forms, each will suffer at the hands of predators conditioned to avoid the other and one will eventually prevail because as soon as it gets the upper hand numerically it will drive the other to extinction. Nonetheless, many species are polymorphic for colour forms that appear to be aposematic. The ladybird beetles (Coccinellidae) furnish a number of particularly striking examples (Hodek, 1973; and see illustration in Ayala, 1978). -

Patterns and Power of Phenotypic Selection in Nature
Articles Patterns and Power of Phenotypic Selection in Nature JOEL G. KINGSOLVER AND DAVID W. PFENNIG Phenotypic selection occurs when individuals with certain characteristics produce more surviving offspring than individuals with other characteristics. Although selection is regarded as the chief engine of evolutionary change, scientists have only recently begun to measure its action in the wild. These studies raise numerous questions: How strong is selection, and do different types of traits experience different patterns of selection? Is selection on traits that affect mating success as strong as selection on traits that affect survival? Does selection tend to favor larger body size, and, if so, what are its consequences? We explore these questions and discuss the pitfalls and future prospects of measuring selection in natural populations. Keywords: adaptive landscape, Cope’s rule, natural selection, rapid evolution, sexual selection henotypic selection occurs when individuals with selection on traits that affect survival stronger than on those Pdifferent characteristics (i.e., different phenotypes) that affect only mating success? In this article, we explore these differ in their survival, fecundity, or mating success. The idea and other questions about the patterns and power of phe- of phenotypic selection traces back to Darwin and Wallace notypic selection in nature. (1858), and selection is widely accepted as the primary cause of adaptive evolution within natural populations.Yet Darwin What is selection, and how does it work? never attempted to measure selection in nature, and in the Selection is the nonrandom differential survival or repro- century following the publication of On the Origin of Species duction of phenotypically different individuals. -

Saving Our Tree HUMAN GENETICS
HIGHLIGHTS EVOLUTION IN BRIEF Saving our tree HUMAN GENETICS Identification of a gene responsible for familial For several years, evolutionary biolo- Wolff–Parkinson–White syndrome. gists interested in gene flow between species have been investigating and Gollob, M. H. et al. N. Engl. J. Med. 344, 1823–1831 (2001) debating a controversial issue: the hor- Wolff–Parkinson–White (WPW) syndrome is an inherited heart izontal exchange of genes between dis- defect associated with arrhythmia, cardiac arrest and sudden death. tantly related species, its extent and Gollob et al. studied two large families with autosomal-dominant possible adaptive significance. WPW syndrome, and used a positional candidate-gene approach to Horizontal gene transfer (HGT) is an identify the causative gene, which encodes a regulatory subunit of important evolutionary force in appears in a cluster of bacterial AMP-activated protein kinase. This finding should shed light on prokaryotes, as their life cycles facilitate sequences, then the vertebrate gene the mechanisms that lead to heart arrhythmias. the exchange of genes between even was probably horizontally transferred distantly related species. However, this from bacteria. Conversely, if all GENETIC NETWORKS is trickier in multicellular organisms, eukaryotic sequences cluster together, in which the fixation of horizontally then HGT has probably not occurred Ordering genes in a flagella pathway by analysis of transferred genes can only occur if as this grouping is most parsimo- expression kinetics from living bacteria. they are transmitted through the germ niously explained by vertebrates hav- Kalir, S. et al. Science 292, 2080–2083 (2001) line. It was therefore surprising when ing acquired the sequence by descent the International Human Genome through other eukaryotes. -

Signals of Predation-Induced Directional and Disruptive Selection in the Threespine Stickleback
Eawag_07048 Evolutionary Ecology Research, 2012, 14: 193–205 Signals of predation-induced directional and disruptive selection in the threespine stickleback Michael Zeller, Kay Lucek, Marcel P. Haesler, Ole Seehausen and Arjun Sivasundar Institute for Ecology and Evolution, University of Bern, Bern, Switzerland and Department of Fish Ecology, Eawag Centre for Ecology, Evolution and Biogeochemistry, Kastanienbaum, Switzerland ABSTRACT Background: Different predation regimes may exert divergent selection pressure on phenotypes and their associated genotypes. Threespine stickleback Gasterosteus aculeatus have a suite of bony structures, which have been shown to be an effective defence against predation and have a well-known genetic basis. Question: Do different predator regimes induce different selective pressures on growth rates and defence phenotypes in threespine stickleback between different habitats across distinct age classes? Hypothesis: In the presence of predation-induced selection, we expect diverging morphological responses between populations experiencing either low or high predation pressure. Study system: Threespine stickleback were sampled from two natural but recently established populations in an invasive range. One site has a high density of fish and insect predators, while at the other site predation pressure is low. Methods: We inferred predator-induced selection on defence traits by comparing the distribution of size classes, defence phenotypes, and an armour-related genotype between dif- ferent age classes in a high and a low predation regime. Results: Under high predation, there are indications of directional selection for faster growth, whereas lateral plate phenotypes and associated genotypes show indications for disruptive selection. Heterozygotes at the Eda-gene have a lower survival rate than either homozygote. Neither pattern is evident in the low predation regime. -

Arise by Chance As the Result of Mutation. They Therefore Suggest
THE EVOLUTION OF DOMINANCE UNDER DISRUPTIVE SELECTION C. A. CLARKE and P. Ni. SHEPPARD Department of Medicine and Department of Zoology, University of Liverpool Received6.iii.59 1.INTRODUCTION INa paper on the effects of disruptive selection, Mather (1955) pointed out that if there are two optimum values for a character and all others are less advantageous or disadvantageous there will be disruptive selection which can lead to the evolution of a polymorphism. Sheppard (1958) argued that where such selection is effective and the change from one optimum value to the other is switched by a single pair of allelomorphs there will be three genotypes but only two advantageous phenotypes. Consequently if dominance were absent initially it would be evolved as a result of the disruptive selection, the heterozygote and one of the homozygotes both coming to resemble one of the two optimum phenotypes (see Ford, 1955, on Tripharna comes). Thoday (1959) has shown by means of an artificial selection experiment that, even when a character is, at the beginning, controlled polygenically (sternopleural chaeta-number in Drosophila) and there is 50 per cent. gene exchange between the "high" and "low" selected sub-popu- lations, a polymorphism can evolve. The most fully understood examples of disruptive selection (other than sex) are provided by instances of Batesian Mimicry, where there are a number of distinct warningly coloured species, acting as models, which are mimicked by the polymorphic forms of a single more edible species. Fisher and Ford (see Ford, 1953) have argued that a suffi- ciently good resemblance between mimic and model is not likely to arise by chance as the result of mutation. -
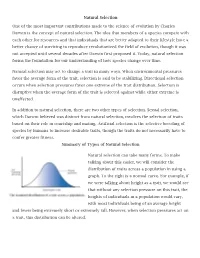
Natural Selection One of the Most Important Contributions Made to The
Natural Selection One of the most important contributions made to the science of evolution by Charles Darwin is the concept of natural selection. The idea that members of a species compete with each other for resources and that individuals that are better adapted to their lifestyle have a better chance of surviving to reproduce revolutionized the field of evolution, though it was not accepted until several decades after Darwin first proposed it. Today, natural selection forms the foundation for our understanding of how species change over time. Natural selection may act to change a trait in many ways. When environmental pressures favor the average form of the trait, selection is said to be stabilizing. Directional selection occurs when selection pressures favor one extreme of the trait distribution. Selection is disruptive when the average form of the trait is selected against while either extreme is unaffected. In addition to natural selection, there are two other types of selection. Sexual selection, which Darwin believed was distinct from natural selection, involves the selection of traits based on their role in courtship and mating. Artificial selection is the selective breeding of species by humans to increase desirable traits, though the traits do not necessarily have to confer greater fitness. Summary of Types of Natural Selection Natural selection can take many forms. To make talking about this easier, we will consider the distribution of traits across a population in using a graph. To the right is a normal curve. For example, if we were talking about height as a trait, we would see that without any selection pressure on this trait, the heights of individuals in a population would vary, with most individuals being of an average height and fewer being extremely short or extremely tall. -

Gene Flow 1 6 James Mallet
Gene Flow 1 6 James Mallet Calton Laboratory, Department of Biology, Univetsity College London, 4 Stephenson Way, London NWI 2HE, UK What is Gene Flow? 'Gene flow' means the movement of genes. In some cases, small fragments of DNA may pass from one individual directly into the germline of another, perhaps transduced by a pathogenic virus or other vector, or deliberately via a human transgenic manipulation. However, this kind of gene flow, known as horizontal gene transfer, is rare. Most of the time, gene flow is caused by the movement or dispersal of whole organisms or genomes from one popula- tion to another. After entering a new population, immigrant genomes may become incorporated due to sexual reproduction or hybridization, and will be gradually broken up by recombination. 'Genotype flow' would therefore be a more logical term to indicate that the whole genome is moving at one time. The term 'gene flow' is used probably because of an implicit belief in abundant recombination, and because most theory is still based on simple single locus models: it does not mean that genes are transferred one at a time. The fact that gene flow is usually caused by genotype flow has important consequences for its measurement, as we shall see. Two Meanings of 'Gene Flow' We are often taught that 'dispersal does not necessarily lead to gene flow'. The term 'gene flow' is then being used in the sense of a final state of the population, i.e. successful establishment of moved genes. This disagrees somewhat with a more straightforward interpretation of gene flow as actual EICAR International 2001. -

Biological Invasions at the Gene Level VIEWPOINT Rémy J
Diversity and Distributions, (Diversity Distrib.) (2004) 10, 159–165 Blackwell Publishing, Ltd. BIODIVERSITY Biological invasions at the gene level VIEWPOINT Rémy J. Petit UMR Biodiversité, Gènes et Ecosystèmes, 69 ABSTRACT Route d’Arcachon, 33612 Cestas Cedex, France Despite several recent contributions of population and evolutionary biology to the rapidly developing field of invasion biology, integration is far from perfect. I argue here that invasion and native status are sometimes best discussed at the level of the gene rather than at the level of the species. This, and the need to consider both natural (e.g. postglacial) and human-induced invasions, suggests that a more integrative view of invasion biology is required. Correspondence: Rémy J. Petit, UMR Key words Biodiversité, Gènes et Ecosystèmes, 69 Route d’Arcachon, 33612 Cestas Cedex, France. Alien, genetic assimilation, gene flow, homogenization, hybridization, introgression, E-mail: [email protected] invasibility, invasiveness, native, Quercus, Spartina. the particular genetic system of plants. Although great attention INTRODUCTION has been paid to the formation of new hybrid taxa, introgression Biological invasions are among the most important driving more often results in hybrid swarms or in ‘genetic pollution’, forces of evolution on our human-dominated planet. According which is best examined at the gene level. Under this perspective, to Myers & Knoll (2001), distinctive features of evolution now translocations of individuals and even movement of alleles include a homogenization of biotas, a proliferation of opportunistic within a species’ range (e.g. following selective sweeps) should species, a decline of biodisparity (the manifest morphological equally be recognized as an important (if often cryptic) and physiological variety of biotas), and increased rates of speci- component of biological invasions. -

Divergence, Gene Flow, and Speciation in Eight Lineages of Trans‐Beringian Birds
Received: 2 May 2019 | Revised: 22 July 2020 | Accepted: 27 July 2020 DOI: 10.1111/mec.15574 ORIGINAL ARTICLE Divergence, gene flow, and speciation in eight lineages of trans-Beringian birds Jessica F. McLaughlin1,2 | Brant C. Faircloth3 | Travis C. Glenn4 | Kevin Winker1 1University of Alaska Museum, Fairbanks, AK, USA Abstract 2Sam Noble Oklahoma Museum of Natural Determining how genetic diversity is structured between populations that span the History, Norman, OK, USA divergence continuum from populations to biological species is key to understanding 3Department of Biological Sciences and Museum of Natural Science, Louisiana State the generation and maintenance of biodiversity. We investigated genetic divergence University, Baton Rouge, LA, USA and gene flow in eight lineages of birds with a trans-Beringian distribution, where 4 Department of Environmental Health Asian and North American populations have likely been split and reunited through Science and Institute of Bioinformatics, University of Georgia, Athens, GA, USA multiple Pleistocene glacial cycles. Our study transects the speciation process, in- cluding eight pairwise comparisons in three orders (ducks, shorebirds and passer- Correspondence Kevin Winker, University of Alaska Museum, ines) at population, subspecies and species levels. Using ultraconserved elements 907 Yukon Drive, Fairbanks, AK 99775, USA. (UCEs), we found that these lineages represent conditions from slightly differentiated Email: [email protected] populations to full biological species. Although allopatric speciation is considered Funding information the predominant mode of divergence in birds, all of our best divergence models in- National Science Foundation, Grant/Award Number: DEB-1242267-1242241-1242260 cluded gene flow, supporting speciation with gene flow as the predominant mode in Beringia.