Chemdis-Mixture: an Online Tool for Analyzing Potential Interaction
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
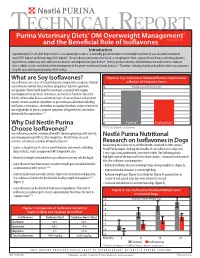
What Are Soy Isoflavones? Figure 2: Soy Isoflavones Reduced Plasma Isoprostanes Soy Isoflavones Are a Class of Natural Bioactive Compounds in Soybeans
TECHNICAL REPORT Purina Veterinary Diets® OM Overweight Management® brand canine dry formula and the Beneficial Role of Isoflavones Introduction Approximately 35% of adult dogs in the U.S. are overweight or obese1. A markedly greater incidence of overweight and obesity was observed in neutered male (59% higher) and female dogs (40% higher)1. Chronic obesity can increase the risk of, or complications from, several chronic diseases including diabetes, hypertension, pulmonary and cardiovascular disease, and degenerative joint disease2. Obesity produces chronic, mild inflammation and increases oxidative stress, which, in turn, contributes to the development of the above-mentioned chronic diseases.3,4 Therefore, reducing obesity and oxidative stress can promote a long life span and improved quality of life in dogs. What are Soy Isoflavones? Figure 2: Soy Isoflavones Reduced Plasma Isoprostanes Soy isoflavones are a class of natural bioactive compounds in soybeans. Natural a Marker of Oxidative Stress soy isoflavones include three chemical compounds: daidzein, genistein, 5 Plasma Isoprostanes (ng/ml) and glycitein. Many health benefits have been associated with regular consumption of soy products. In humans, soy has been found to reduce the 4 risk of cardiovascular disease and certain types of cancer (breast and prostate cancer); relieve a number of problems in post menopausal women including 3 hot flashes, osteoporosis, and decline in cognitive function; reduce cholesterol and triglycerides in plasma; improve symptoms of hypertension; and reduce 2 abdominal fat accumulation.5,6,7 1 Why Did Nestlé Purina 0 Control Isoflavones* Choose Isoflavones? *P<0.01 for Control vs. Isoflavones Soy isoflavones provide a number of benefits for managing dogs with obesity, and reducing rebound effects after weight loss. -

Applications of in Silico Methods to Analyze the Toxicity and Estrogen T Receptor-Mediated Properties of Plant-Derived Phytochemicals ∗ K
Food and Chemical Toxicology 125 (2019) 361–369 Contents lists available at ScienceDirect Food and Chemical Toxicology journal homepage: www.elsevier.com/locate/foodchemtox Applications of in silico methods to analyze the toxicity and estrogen T receptor-mediated properties of plant-derived phytochemicals ∗ K. Kranthi Kumara, P. Yugandharb, B. Uma Devia, T. Siva Kumara, N. Savithrammab, P. Neerajaa, a Department of Zoology, Sri Venkateswara University, Tirupati, 517502, India b Department of Botany, Sri Venkateswara University, Tirupati, 517502, India ARTICLE INFO ABSTRACT Keywords: A myriad of phytochemicals may have potential to lead toxicity and endocrine disruption effects by interfering Phytochemicals with nuclear hormone receptors. In this examination, the toxicity and estrogen receptor−binding abilities of a QSAR modeling set of 2826 phytochemicals were evaluated. The endpoints mutagenicity, carcinogenicity (both CAESAR and ISS Toxicity models), developmental toxicity, skin sensitization and estrogen receptor relative binding affinity (ER_RBA) Nuclear hormone receptor binding were studied using the VEGA QSAR modeling package. Alongside the predictions, models were providing pos- Self−Organizing maps sible information for applicability domains and most similar compounds as similarity sets from their training Clustering and classification schemes sets. This information was subjected to perform the clustering and classification of chemicals using Self−Organizing Maps. The identified clusters and their respective indicators were considered as potential hotspot structures for the specified data set analysis. Molecular screening interpretations of models wereex- hibited accurate predictions. Moreover, the indication sets were defined significant clusters and cluster in- dicators with probable prediction labels (precision). Accordingly, developed QSAR models showed good pre- dictive abilities and robustness, which observed from applicability domains, representation spaces, clustering and classification schemes. -

Daidzein and Genistein Content of Cereals
European Journal of Clinical Nutrition (2002) 56, 961–966 ß 2002 Nature Publishing Group All rights reserved 0954–3007/02 $25.00 www.nature.com/ejcn ORIGINAL COMMUNICATION Daidzein and genistein content of cereals J Liggins1, A Mulligan1,2, S Runswick1 and SA Bingham1,2* 1Medical Research Council Dunn Human Nutrition Unit, Hills Road, Cambridge, UK; and 2European Prospective Investigation of Cancer, University of Cambridge, Cambridge, UK Objective: To analyse 75 cereals and three soy flours commonly eaten in Europe for the phytoestrogens daidzein and genistein. Design: The phytoestrogens daidzein and genistein were extracted from dried foods, and the two isoflavones quantified after hydrolytic removal of any conjugated carbohydrate. Completeness of extraction and any procedural losses of the isoflavones 0 0 were accounted for using synthetic daidzin (7-O-glucosyl-4 -hydroxyisoflavone) and genistin (7-O-glucosyl-4 5-dihydroxyiso- flavone) as internal standards. Setting: Foods from the Cambridge UK area were purchased, prepared for eating, which included cooking if necessary, and freeze dried. Three stock soy flours were also analysed. Results: Eighteen of the foods assayed contained trace or no detectable daidzein or genistein. The soy flours were rich sources, containing 1639 – 2117 mg=kg. The concentration of the two isoflavones in the remaining foods ranged from 33 to 11 873 mg=kg. Conclusion: These analyses will supply useful information to investigators determining the intake of phytoestrogens in cereal products in order to relate intakes to potential biological activities. Sponsorship: This work was supported by the United Kingdom Medical Research Council, Ministry of Agriculture Fisheries and Food (contract FS2034) and the United States of America Army (contract DAMD 17-97-1-7028). -

Mindy Goldman, MD Clinical Professor Dept
Managing Menopause Medically and Naturally Mindy Goldman, MD Clinical Professor Dept. of Ob/Gyn and Reproductive Sciences Director, Women’s Cancer Care Program, UCSF Breast Care Center and Women’s Health University of California, San Francisco I have nothing to disclose –Mindy Goldman, MD CASE STUDY 50 yr. old G2P2 peri-menopausal woman presents with complaints of significant night sweats interfering with her ability to sleep. She has mild hot flashes during the day. She has never had a bone mineral density test but her mother had a hip fracture at age 62 due to osteoporosis. Her 46 yr. old sister was diagnosed with breast cancer at age 43, treated with lumpectomy and radiation and currently is doing well. There is no other family history of cancer. Questions 1. Would you offer her MHT? 2. If yes, how long would you continue it? 3. If no, what would you offer for alternative treatments? 4. Would your treatment differ if you knew she had underlying heart disease? Is it safe? How long can I take it? What about Mymy Bones?bones? Will it protect my heart? MHT - 2015 What about my brain? Will I get breast cancer? What about my hot flashes? Menopausal Symptoms Hot flashes Night sweats Sleep disturbances Vaginal dryness/Sexual dysfunction Mood disturbances How to Treat Menopausal Symptoms Hormone therapy Alternatives to hormones Complementary and Integrative Techniques Prior to Women’s Health Initiative Hormone therapy primary treatment of menopausal hot flashes Few women would continue hormones past one year By 1990’s well known -

Simultaneous Determination of Daidzein, Genistein and Formononetin in Coffee by Capillary Zone Electrophoresis
separations Article Simultaneous Determination of Daidzein, Genistein and Formononetin in Coffee by Capillary Zone Electrophoresis Feng Luan *, Li Li Tang, Xuan Xuan Chen and Hui Tao Liu College of Chemistry and Chemical Engineering, Yantai University, Yantai 264005, China; [email protected] (L.L.T.); [email protected] (X.X.C.); [email protected] (H.T.L.) * Correspondence: fl[email protected]; Tel.: +86-535-6902063 Academic Editor: Doo Soo Chung Received: 29 October 2016; Accepted: 20 December 2016; Published: 1 January 2017 Abstract: Coffee is a favorite and beverage in Western countries that is consumed daily. In the present study, capillary zone electrophoresis (CE) was applied for the separation and quantification of three isoflavones including daidzein, genistein and formononetin in coffee. Extraction of isoflavones from the coffee sample was carried out by extraction and purification process using ether after the acid hydrolysis with the antioxidant butylated hydroxy-toluene (BHT). The experimental conditions of the CE separation method were: 20 mmol/L Na2HPO4 buffer solution, 25 kV applied voltage, 3 s hydrodynamic injection at 30 mbar, and UV detection at 254 nm. The results show that the three compounds can be tested within 10 min with a linearity of 0.5–50 µg/mL for all three compounds. The limits of detection were 0.0642, 0.134, and 0.0825 µg/mL for daidzein, formononetin and genistein, respectively. The corresponding average recovery was 99.39% (Relative Standard Detection (RSD) = 1.76%), 98.71% (RSD = 2.11%) and 97.37% (RSD = 3.74%). Keywords: capillary zone electrophoresis (CE); daidzein; genistein; formononetin; acid hydrolysis 1. -

Contemporary Approaches for Managing Menopause Symptoms
Menopause 2017 Guidelines: Looking at Special Populations and Building on Existing Practice Patricia Geraghty, MSN, FNP-BC, WHNP Disclosures • Speaker Bureau • AbbVie • Therapeutics MD • Advisory Board • AbbVie (endometriosis) • Sharecare Inc. • Procedure Proctor • Bayer (Contraception) • Off label discussion will be included and identified in this discussion. Objectives • Identify the benefits and risks of estrogen, progesterone, and non-hormonal pharmacological management of menopause symptoms based on dosage, route of administration, pharmacokinetics, treatment population and duration of use. • Develop treatment strategies for special populations including premature ovarian insufficiency, prolonged symptoms, and women who are not candidates for estrogen. • Incorporate the NAMS 2017 Position Statement into patient counseling on the timeline of menopause symptoms and the termination of hormone therapy. Lifespan Lifespan Median 34 322 BC Aristotle describes transition Ages the Through Menopause yr JAMA. JAMA. Investigators. Initiative Health the Women's for Group Writing 2002;288(3):321 - 333. doi:10.1001/jama.288.3.321 Treated with plants s/a 1800’s cohosh, cannabis, opium 1821 French physician DeGardanne 53 lifespan lifespan calls it “Menopause” Adult - 1900’s Deficiency disease 55 1942 Premarin” copyright yr 1960’s “Forever Feminine” by Robert Wilson MD 1980’s-1990’s “Politics of Menopause” by Frances McCrea 2002 WHI- First large randomized control trial “My periods are different. Is This Menopause?” • Random cycling, sometimes early, sometimes late. • Flow variable with prodromal spotting, a long taper, or stopping and starting. • She has a day of very heavy flow. • 77% have duration 10+ days, heavy bleeding 3+ days, spotting 6+ days Paramsothy P, et al. BJOG. 2014 Nov;121(12):1564-73. -

Inhibitory Effect of Genistein and Daidzein on Ovarian Cancer Cell Growth
ANTICANCER RESEARCH 24: 795-800 (2004) Inhibitory Effect of Genistein and Daidzein on Ovarian Cancer Cell Growth CICEK GERCEL-TAYLOR, ANNA K. FEITELSON and DOUGLAS D. TAYLOR Department of Obstetrics, Gynecology and Women’s Health, University of Louisville, School of Medicine, Louisville, KY 40202, U.S.A. Abstract. Background: Survival from ovarian cancer has not Genistein, a soy isoflavanoid, has been intensely studied changed significantly in the past twenty years requiring in relation to breast cancer. Interest first arose upon development of additional treatment protocols. We studied the discovery of the vast difference in breast cancer rates in effect of genistein and daidzein on ovarian cancer cell growth. Asia versus Western countries (1). Large dietary differences Materials and Methods: Five ovarian cancer cell lines from Stage exist, especially in genistein consumption, as the average IIIC disease were evaluated. Sulforhodamine B and colony Asian intake is 20-80 mg/day whereas the average US intake formation assays were used to analyze growth inhibitory effects of is only 1-3 mg (2,3) The dietary and disease discrepancy genistein and daidzein alone and with cisplatin, paclitaxel or prompted further study into the chemopreventive and topotecan. Apoptosis induction was studied by determining potentially therapeutic properties of genistein. caspase-3 activity. Results: Inhibition of growth (50-80%), colony Genistein has been found to inhibit cell proliferation, formation and colony size was seen at 144 Ìm of genistein, 0-23% oncogenesis and clonogenic ability in animal and human reduction was demonstrated at 9 Ìm. At 144 Ìm, the colony size cells (3-5). Several studies have been performed to evaluate was inhibited >75%; at 9 Ìm 4/5 cell lines had >50% reduction. -

Daidzein and Breast Cancer Risk in Later Years FACT SHEET on the PHYTOESTROGEN DAIDZEIN
BREAST CANCER & THE ENVIRONMENT RESEARCH CENTERS Early Life Exposure to the Phytoestrogen Daidzein and Breast Cancer Risk in Later Years FACT SHEET on the PHYTOESTROGEN DAIDZEIN Abstract Daidzein is a phytoestrogen (estrogen-like chemical compound present in plants) that binds to estrogen receptors and has both weak estrogenic and weak anti-estrogenic effects. There are three major classes of phytoestrogens that have estrogen-like actions in the human body. They are lignans, isoflavones, and coumestans. Daidzein is an isoflavone. Exposure to daidzein occurs principally through foods made with soybeans and soy protein. In a proportion of the population, daidzein is metabolized by intestinal bacteria to produce equol and O-DMA, metabolites that are more estrogenic than daidzein. Daidzein can cross the placenta and has been found in breast milk. It is unknown whether daidzein influences early onset of puberty in girls. Exposure to daidzein can be measured using a blood or urine test; however levels vary widely in each person due to considerable variability in the metabolism of daidzein. In vitro and in vivo studies have found that daidzein stimulates the growth of estrogen-sensitive breast cancer cells. Epidemiologic studies have found conflicting evidence; some studies have found an association between soy exposure and decreased breast cancer risk while others have found no association. Some epidemiological evidence indicates that soy intake may be more protective when the exposure occurs prior to puberty. More research needs to be conducted on the association between breast cancer risk and daidzein specifically before conclusions can be drawn. The International Agency for Research on Cancer (IARC) has not determined whether phytoestrogens are carcinogenic to humans. -

Dr. Duke's Phytochemical and Ethnobotanical Databases List of Chemicals for Dry Mouth / Xerostomia
Dr. Duke's Phytochemical and Ethnobotanical Databases List of Chemicals for Dry Mouth / Xerostomia Chemical Activity Count (+)-CATECHIN 2 (+)-EPIPINORESINOL 1 (-)-ANABASINE 1 (-)-EPICATECHIN 2 (-)-EPIGALLOCATECHIN 2 (-)-EPIGALLOCATECHIN-GALLATE 2 (Z)-1,3-BIS(4-HYDROXYPHENYL)-1,4-PENTADIENE 1 1,8-CINEOLE 2 10-METHOXYCAMPTOTHECIN 1 16-HYDROXY-4,4,10,13-TETRAMETHYL-17-(4-METHYL-PENTYL)-HEXADECAHYDRO- 1 CYCLOPENTA[A]PHENANTHREN-3-ONE 2,3-DIHYDROXYBENZOIC-ACID 1 3'-O-METHYL-CATECHIN 1 3-ACETYLACONITINE 1 3-O-METHYL-(+)-CATECHIN 1 4-O-METHYL-GLUCURONOXYLAN 1 5,7-DIHYDROXY-2-METHYLCHROMONE-8-C-BETA-GLUCOPYRANOSIDE 1 5-HYDROXYTRYPTAMINE 1 5-HYDROXYTRYPTOPHAN 1 6-METHOXY-BENZOLINONE 1 ACEMANNAN 1 ACETYL-CHOLINE 1 ACONITINE 2 ADENOSINE 2 AFFINISINE 1 AGRIMONIIN 1 ALANTOLACTONE 2 ALKANNIN 1 Chemical Activity Count ALLANTOIN 1 ALLICIN 2 ALLIIN 2 ALLOISOPTEROPODINE 1 ALLOPTEROPODINE 1 ALLOPURINOL 1 ALPHA-LINOLENIC-ACID 1 ALPHA-TERPINEOL 1 ALPHA-TOCOPHEROL 2 AMAROGENTIN 1 AMELLIN 1 ANABASINE 1 ANDROMEDOTOXIN 1 ANETHOLE 1 ANTHOCYANIDINS 1 ANTHOCYANINS 1 ANTHOCYANOSIDE 1 APIGENIN 1 APOMORPHINE 1 ARABINO-3,6-GALACTAN-PROTEIN 1 ARABINOGALACTAN 1 ARACHIDONIC-ACID 1 ARCTIGENIN 2 ARECOLINE 1 ARGLABRIN 1 ARISTOLOCHIC-ACID 1 ARISTOLOCHIC-ACID-I 1 2 Chemical Activity Count ARMILLARIEN-A 1 ARTEMISININ 1 ASCORBIC-ACID 4 ASTRAGALAN-I 1 ASTRAGALAN-II 1 ASTRAGALAN-III 1 ASTRAGALIN 1 AURICULOSIDE 1 BAICALEIN 1 BAICALIN 1 BAKUCHIOL 1 BENZALDEHYDE 1 BERBAMINE 1 BERBERASTINE 3 BERBERINE 3 BERBERINE-CHLORIDE 1 BERBERINE-IODIDE 1 BERBERINE-SULFATE 1 BETA-AMYRIN-PALMITATE -

The Influence of Plant Isoflavones Daidzein and Equol on Female
pharmaceuticals Review The Influence of Plant Isoflavones Daidzein and Equol on Female Reproductive Processes Alexander V. Sirotkin 1,* , Saleh Hamad Alwasel 2 and Abdel Halim Harrath 2 1 Department of Zoology and Anthropology, Constantine the Philosopher University in Nitra, 949 01 Nitra, Slovakia 2 Department of Zoology, College of Science, King Saud University, Riyadh 12372, Saudi Arabia; [email protected] (S.H.A.); [email protected] (A.H.H.) * Correspondence: [email protected]; Tel.: +421-903561120 Abstract: In this review, we explore the current literature on the influence of the plant isoflavone daidzein and its metabolite equol on animal and human physiological processes, with an emphasis on female reproduction including ovarian functions (the ovarian cycle; follicullo- and oogenesis), fundamental ovarian-cell functions (viability, proliferation, and apoptosis), the pituitary and ovarian endocrine regulators of these functions, and the possible intracellular mechanisms of daidzein action. Furthermore, we discuss the applicability of daidzein for the control of animal and human female reproductive processes, and how to make this application more efficient. The existing literature demonstrates the influence of daidzein and its metabolite equol on various nonreproductive and reproductive processes and their disorders. Daidzein and equol can both up- and downregulate the ovarian reception of gonadotropins, healthy and cancerous ovarian-cell proliferation, apoptosis, viability, ovarian growth, follicullo- and oogenesis, and follicular atresia. These effects could be mediated by daidzein and equol on hormone production and reception, reactive oxygen species, and intracellular regulators of proliferation and apoptosis. Both the stimulatory and the inhibitory Citation: Sirotkin, A.V.; Alwasel, effects of daidzein and equol could be useful for reproductive stimulation, the prevention and S.H.; Harrath, A.H. -

Washington, Dc September 21-24, 2011
ABSTRACT BOOK 22ND ANNUAL MEETING Gaylord National on the Potomac WASHINGTON, DC SEPTEMBER 21-24, 2011 Also includes: • Disclosures for all presenters • Speakers’ learning objectives and recommended reading What happens in DC doesn’t have to stay in DC The webcast of the 2011 NAMS Annual Meeting lets you take advantage of the meeting’s educational riches year-round, at your convenience. If you miss part of the meeting or just want to revisit some sessions, this free on-demand webcast is your answer. The webcast will capture all plenary sessions as well as the Pre-Meeting Symposium and let you select individual presentations for targeted viewing. It will be freely available to all through September 15, 2012. It’s a great way to reinforce your learning at your leisure and according to your schedule. The free webcast will be posted soon after the meeting on the NAMS website at: www.menopause.org/meetings/webcast.aspx Stay tuned for webcast launch announcements from NAMS via email blast and Facebook and Twitter postings. Photos used with permission from Microsoft 270 Webcast2011_PreLaunch_Sidebar.indd 1 8/16/11 9:43 AM Contents Key to Abstracts . 4 Invited Speakers’ Abstracts & Learning Objectives . 7 Scientific Session Speakers’ Abstracts . 32 Basic Science Poster Presentations . 40 Clinical Poster Presentations . 44 Disclosure Statement . 67 Key to Disclosures . 68 Disclosures . 69 Invited Speakers’ Recommended Reading . 73 Call for Abstracts 2012 Annual Meeting . Inside Back Cover 3 Key to Abstracts Invited Speakers’ Abstracts Scientific Session Speakers’ Abstracts Speakers Page # Speakers Page # David F . Archer, MD, NCMP . 19 Susan E . Appt, DVM . -

The Vascular Effects of Isolated Isoflavones—A Focus on The
biology Review The Vascular Effects of Isolated Isoflavones—A Focus on the Determinants of Blood Pressure Regulation Henrique Silva 1,2 1 Informetrics Research Group, Ton Duc Thang University, Ho Chi Minh City 758307, Vietnam; [email protected] 2 Faculty of Pharmacy, Ton Duc Thang University, Ho Chi Minh City 758307, Vietnam Simple Summary: Isoflavones are naturally-occurring phytoestrogens, highly prevalent in soybeans, and known to improve cardiovascular health in populations with a high isoflavone dietary intake. Most clinical studies have assessed the impact of dietary intake or supplementation with mixtures of isoflavones, with few studies dedicated to the effects of isolated compounds (i.e., genistein, daidzein, glycitein, formononetin, biochanin A, and equol). This paper reviews the main actions of isolated isoflavones on the vasculature, with particular focus on the determinants of blood pressure regulation. Isoflavones evoke relaxation of different vascular beds by acting on several signaling pathways in the endothelium, where they potentiate the release of important vasorelaxant mediators, and in vascular smooth muscle cells, where relaxation is attained mainly through hyperpolarization. Some of these effects are attributed to their ability to modulate estrogen receptors. These vascular effects occur at plasma concentrations in the micromolar range, attained only through dietary supplementation. This paper highlights isolated isoflavones as potentially suitable alternatives to soy-based foodstuffs and supplements and which could enlarge the current therapeutic arsenal. Abstract: Isoflavones are phytoestrogen compounds with important biological activities, including improvement of cardiovascular health. This activity is most evident in populations with a high isoflavone dietary intake, essentially from soybean-based products. The major isoflavones known to Citation: Silva, H.