Homework Solution
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

CCA One Care Options Formulary
Commonwealth Care Alliance One Care Plan (Medicare-Medicaid Plan) 2021 List of Covered Drugs (Formulary) 30 Winter Street • Boston, MA 02108 PLEASE READ: THIS DOCUMENT CONTAINS INFORMATION ABOUT THE DRUGS WE COVER IN THIS PLAN For more recent information or other questions, contact Commonwealth Care Alliance Member Services at 1-866-610-2273 (TTY: call MassRelay at 711), 8 a.m. – 8 p.m., 7 days a week, or visit www.commonwealthonecare.org H0137_CF2021 Approved Formulary: ID 00021588 • Version 13 • Updated on 08/01/2021 One Care Plan | 2021 List of Covered Drugs (Formulary) Introduction This document is called the List of Covered Drugs (also known as the Drug List). It tells you which prescription drugs, over-the-counter drugs and items are covered by Commonwealth Care Alliance. The Drug List also tells you if there are any special rules or restrictions on any drugs covered by One Care. Key terms and their definitions appear in the last chapter of the Member Handbook. Table of Contents A. Disclaimers ........................................................................................................................ 4 B. Frequently Asked Questions (FAQ) .................................................................................. 5 What prescription drugs are on the List of Covered Drugs? (We call the List of Covered Drugs the “Drug List” for short.) ................................................................... 5 B2. Does the Drug List ever change? ............................................................................... 5 B3. What happens when there is a change to the Drug List? ........................................... 6 B4. Are there any restrictions or limits on drug coverage or any required actions to take to get certain drugs? .................................................................................................. 7 B5. How will you know if the drug you want has limitations or if there are required actions to take to get the drug? ................................................................................. -

Direct Sensing and Discrimination Among Ubiquitin and Ubiquitin Chains Using Solid-State Nanopores
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector 2340 Biophysical Journal Volume 108 May 2015 2340–2349 Article Direct Sensing and Discrimination among Ubiquitin and Ubiquitin Chains Using Solid-State Nanopores Iftach Nir,1 Diana Huttner,1 and Amit Meller1,* 1Department of Biomedical Engineering, The Technion—Israel Institute of Technology, Haifa, Israel ABSTRACT Nanopore sensing involves an electrophoretic transport of analytes through a nanoscale pore, permitting label- free sensing at the single-molecule level. However, to date, the detection of individual small proteins has been challenging, primarily due to the poor signal/noise ratio that these molecules produce during passage through the pore. Here, we show that fine adjustment of the buffer pH, close to the isoelectric point, can be used to slow down the translocation speed of the analytes, hence permitting sensing and characterization of small globular proteins. Ubiquitin (Ub) is a small protein of 8.5 kDa, which is well conserved in all eukaryotes. Ub conjugates to proteins as a posttranslational modification called ubiquiti- nation. The immense diversity of Ub substrates, as well as the complexity of Ub modification types and the numerous physio- logical consequences of these modifications, make Ub and Ub chains an interesting and challenging subject of study. The ability to detect Ub and to identify Ub linkage type at the single-molecule level may provide a novel tool for investigation in the Ub field. This is especially adequate because, for most ubiquitinated substrates, Ub modifies only a few molecules in the cell at a given time. -
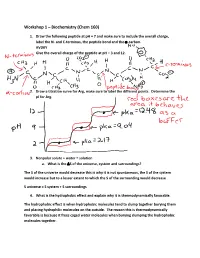
Workshop 1 – Biochemistry (Chem 160)
Workshop 1 – Biochemistry (Chem 160) 1. Draw the following peptide at pH = 7 and make sure to include the overall charge, label the N- and C-terminus, the peptide bond and the -carbon. AVDKY Give the overall charge of the peptide at pH = 3 and 12. 2. Draw a titration curve for Arg, make sure to label the different points. Determine the pI for Arg. 3. Nonpolar solute + water = solution a. What is the S of the universe, system and surroundings? The S of the universe would decrease this is why it is not spontaneous, the S of the system would increase but to a lesser extent to which the S of the surrounding would decrease S universe = S system + S surroundings 4. What is the hydrophobic effect and explain why it is thermodynamically favorable. The hydrophobic effect is when hydrophobic molecules tend to clump together burying them and placing hydrophilic molecules on the outside. The reason this is thermodynamically favorable is because it frees caged water molecules when burying clumping the hydrophobic molecules together. 5. Urea dissolves very readily in water, but the solution becomes very cold as the urea dissolves. How is this possible? Urea dissolves in water because when dissolving there is a net increase in entropy of the universe. The heat exchange, getting colder only reflects the enthalpy (H) component of the total energy change. The entropy change is high enough to offset the enthalpy component and to add up to an overall -G 6. A mutation that changes an alanine residue in the interior of a protein to valine is found to lead to a loss of activity. -

Amino Acid Recognition by Aminoacyl-Trna Synthetases
www.nature.com/scientificreports OPEN The structural basis of the genetic code: amino acid recognition by aminoacyl‑tRNA synthetases Florian Kaiser1,2,4*, Sarah Krautwurst3,4, Sebastian Salentin1, V. Joachim Haupt1,2, Christoph Leberecht3, Sebastian Bittrich3, Dirk Labudde3 & Michael Schroeder1 Storage and directed transfer of information is the key requirement for the development of life. Yet any information stored on our genes is useless without its correct interpretation. The genetic code defnes the rule set to decode this information. Aminoacyl-tRNA synthetases are at the heart of this process. We extensively characterize how these enzymes distinguish all natural amino acids based on the computational analysis of crystallographic structure data. The results of this meta-analysis show that the correct read-out of genetic information is a delicate interplay between the composition of the binding site, non-covalent interactions, error correction mechanisms, and steric efects. One of the most profound open questions in biology is how the genetic code was established. While proteins are encoded by nucleic acid blueprints, decoding this information in turn requires proteins. Te emergence of this self-referencing system poses a chicken-or-egg dilemma and its origin is still heavily debated 1,2. Aminoacyl-tRNA synthetases (aaRSs) implement the correct assignment of amino acids to their codons and are thus inherently connected to the emergence of genetic coding. Tese enzymes link tRNA molecules with their amino acid cargo and are consequently vital for protein biosynthesis. Beside the correct recognition of tRNA features3, highly specifc non-covalent interactions in the binding sites of aaRSs are required to correctly detect the designated amino acid4–7 and to prevent errors in biosynthesis5,8. -

Amino Acid Chemistry
Handout 4 Amino Acid and Protein Chemistry ANSC 619 PHYSIOLOGICAL CHEMISTRY OF LIVESTOCK SPECIES Amino Acid Chemistry I. Chemistry of amino acids A. General amino acid structure + HN3- 1. All amino acids are carboxylic acids, i.e., they have a –COOH group at the #1 carbon. 2. All amino acids contain an amino group at the #2 carbon (may amino acids have a second amino group). 3. All amino acids are zwitterions – they contain both positive and negative charges at physiological pH. II. Essential and nonessential amino acids A. Nonessential amino acids: can make the carbon skeleton 1. From glycolysis. 2. From the TCA cycle. B. Nonessential if it can be made from an essential amino acid. 1. Amino acid "sparing". 2. May still be essential under some conditions. C. Essential amino acids 1. Branched chain amino acids (isoleucine, leucine and valine) 2. Lysine 3. Methionine 4. Phenyalanine 5. Threonine 6. Tryptophan 1 Handout 4 Amino Acid and Protein Chemistry D. Essential during rapid growth or for optimal health 1. Arginine 2. Histidine E. Nonessential amino acids 1. Alanine (from pyruvate) 2. Aspartate, asparagine (from oxaloacetate) 3. Cysteine (from serine and methionine) 4. Glutamate, glutamine (from α-ketoglutarate) 5. Glycine (from serine) 6. Proline (from glutamate) 7. Serine (from 3-phosphoglycerate) 8. Tyrosine (from phenylalanine) E. Nonessential and not required for protein synthesis 1. Hydroxyproline (made postranslationally from proline) 2. Hydroxylysine (made postranslationally from lysine) III. Acidic, basic, polar, and hydrophobic amino acids A. Acidic amino acids: amino acids that can donate a hydrogen ion (proton) and thereby decrease pH in an aqueous solution 1. -

Marginal Protein Stability Drives Subcellular Proteome Isoelectric Point
Marginal protein stability drives subcellular proteome isoelectric point Kaiser Loella,b and Vikas Nandaa,b,1 aCenter for Advanced Biotechnology and Medicine, Rutgers University, Piscataway, NJ 08854; and bDepartment of Biochemistry and Molecular Biology, Robert Wood Johnson Medical School, Rutgers University, Piscataway, NJ 08854 Edited by David Baker, University of Washington, Seattle, WA, and approved October 3, 2018 (received for review May 26, 2018) There exists a positive correlation between the pH of subcellular matching subcellular pH. Such selection could apply broadly compartments and the median isoelectric point (pI) for the across many proteins, resulting in proteome-wide effects (12). associated proteomes. Proteins in the human lysosome—a highly However, rather than exhibiting high stability under physiolog- acidic compartment in the cell—have a median pI of ∼6.5, whereas ical conditions, the majority of proteins are marginally stable, with proteins in the more basic mitochondria have a median pI of ∼8.0. free energy differences of only 5 kcal/mol to 15 kcal/mol between Proposed mechanisms reflect potential adaptations to pH. For ex- the folded and unfolded states (16). Neutral evolution theory ample, enzyme active site general acid/base residue pKs are likely posits most diversity can be explained by the accumulation of evolved to match environmental pH. However, such effects would random mutations that have minimal impact on fitness (17). be limited to a few residues on specific proteins, and might not Models of protein evolution demonstrate that proteome-wide affect the proteome at large. A protein model that considers res- marginal stability can be understood as neutral, rather than pos- idue burial upon folding recapitulates the correlation between itive selection for instability (18, 19). -

Amino Acids Amino Acids
Amino Acids Amino Acids What Are Amino Acids? Essential Amino Acids Non Essential Amino Acids Amino acids are the building blocks of proteins; proteins are made of amino acids. Isoleucine Arginine (conditional) When you ingest a protein your body breaks it down into the individual aminos, Leucine Glutamine (conditional) reorders them, re-folds them, and turns them into whatever is needed by the body at Lysine Tyrosine (conditional) that time. From only 20 amino acids, the body is able to make thousands of unique proteins with different functions. Methionine Cysteine (conditional) Phenylalanine Glycine (conditional) Threonine Proline (conditional) Did You Know? Tryptophan Serine (conditional) Valine Ornithine (conditional) There are 20 different types of amino acids that can be combined to make a protein. Each protein consists of 50 to 2,000 amino acids that are connected together in a specific Histidine* Alanine sequence. The sequence of the amino acids determines each protein’s unique structure Asparagine and its specific function in the body. Asparate Popular Amino Acid Supplements How Do They Benefit Our Health? Acetyl L- Carnitine: As part of its role in supporting L-Lysine: L-Lysine, an essential amino acid, is mental function, Acetyl L-Carnitine may help needed to support proper growth and bone Proteins (amino acids) are needed by your body to maintain muscles, bones, blood, as support memory, attention span and mental development. It can also support immune function. well as create enzymes, neurotransmitters and antibodies, as well as transport and performance. store molecules. N-Acetyl Cysteine: N-Acetyl Cysteine (NAC) is a L-Arginine: L-Arginine is a nonessential amino acid form of the amino acid cysteine. -

Solutions to 7.012 Problem Set 1
MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Solutions to 7.012 Problem Set 1 Question 1 Bob, a student taking 7.012, looks at a long-standing puddle outside his dorm window. Curious as to what was growing in the cloudy water, he takes a sample to his TA, Brad Student. He wanted to know whether the organisms in the sample were prokaryotic or eukaryotic. a) Give an example of a prokaryotic and a eukaryotic organism. Prokaryotic: Eukaryotic: All bacteria Yeast, fungi, any animial or plant b) Using a light microscope, how could he tell the difference between a prokaryotic organism and a eukaryotic one? The resolution of the light microscope would allow you to see if the cell had a true nucleus or organelles. A cell with a true nucleus and organelles would be eukaryotic. You could also determine size, but that may not be sufficient to establish whether a cell is prokaryotic or eukaryotic. c) What additional differences exist between prokaryotic and eukaryotic organisms? Any answer from above also fine here. In addition, prokaryotic and eukaryotic organisms differ at the DNA level. Eukaryotes have more complex genomes than prokaryotes do. Question 2 A new startup company hires you to help with their product development. Your task is to find a protein that interacts with a polysaccharide. a) You find a large protein that has a single binding site for the polysaccharide cellulose. Which amino acids might you expect to find in the binding pocket of the protein? What is the strongest type of interaction possible between these amino acids and the cellulose? Cellulose is a polymer of glucose and as such has many free hydroxyl groups. -

List of Toxic Chemicals Within the Glycol Ethers Category
United States Office of Environmental Revised December 2000 Environmental Protection Information EPA 745-R-00-004 Agency Washington, DC 20460 TOXICS RELEASE INVENTORY List of Toxic Chemicals within the Glycol Ethers Category Section 313 of the Emergency Planning and Community Right-to-Know Act (EPCRA) requires certain facilities manufacturing, processing, or otherwise using listed toxic chemicals to report their environmental releases of such chemicals annually. Beginning with the 1991 reporting year, such facilities also must report pollution prevention and recycling data for such chemicals, pursuant to section 6607 of the Pollution Prevention Act, 42 U.S.C. 13106. When enacted, EPCRA section 313 established an initial list of toxic chemicals that was comprised of more than 300 chemicals and 20 chemical categories. EPCRA section 313(d) authorizes EPA to add chemicals to or delete chemicals from the list, and sets forth criteria for these actions. CONTENTS Section 1. Introduction ...................................................... 3 Section 2. CAS Number List of Some Chemicals within the Glycol Ethers Category ........ 6 Section 3. CAS Number List of Some Mixtures That Contain Glycol Ethers within the Category .............................................. 185 Section 4. CAS Number List of Some Oligomeric or Polymeric Chemicals That Might Contain Glycol Ether Components within the Category .......................... 187 FOREWORD This document is an updated version of the previous document, EPA 745-R-99-006, June 1999. This version has the following updates: • The titles to Table 1 on page 6, Table 2 on page 185, and Table 3 on 187 are modified; and • The CAS number of second listing in Table 3 (Poly(oxy-1,2-ethanediyl), .alpha.- (phenylsulfonyl)-.omega.-methoxy-) on page 187 is changed from 7664-41-7 to 67584-43-4. -

Nucleotide Base Coding and Am1ino Acid Replacemients in Proteins* by Emil L
VOL. 48, 1962 BIOCHEMISTRY: E. L. SAIITH 677 18 Britten, R. J., and R. B. Roberts, Science, 131, 32 (1960). '9 Crestfield, A. M., K. C. Smith, and F. WV. Allen, J. Biol. Chem., 216, 185 (1955). 20 Gamow, G., Nature, 173, 318 (1954). 21 Brenner, S., these PROCEEDINGS, 43, 687 (1957). 22 Nirenberg, M. WV., J. H. Matthaei, and 0. WV. Jones, unpublished data. 23 Crick, F. H. C., L. Barnett, S. Brenner, and R. J. Watts-Tobin, Nature, 192, 1227 (1961). 24 Levene, P. A., and R. S. Tipson, J. Biol. Ch-nn., 111, 313 (1935). 25 Gierer, A., and K. W. Mundry, Nature, 182, 1437 (1958). 2' Tsugita, A., and H. Fraenkel-Conrat, J. Mllot. Biol., in press. 27 Tsugita, A., and H. Fraenkel-Conrat, personal communication. 28 Wittmann, H. G., Naturwissenschaften, 48, 729 (1961). 29 Freese, E., in Structure and Function of Genetic Elements, Brookhaven Symposia in Biology, no. 12 (1959), p. 63. NUCLEOTIDE BASE CODING AND AM1INO ACID REPLACEMIENTS IN PROTEINS* BY EMIL L. SMITHt LABORATORY FOR STUDY OF HEREDITARY AND METABOLIC DISORDERS AND THE DEPARTMENTS OF BIOLOGICAL CHEMISTRY AND MEDICINE, UNIVERSITY OF UTAH COLLEGE OF MEDICINE Communicated by Severo Ochoa, February 14, 1962 The problem of which bases of messenger or template RNA' specify the coding of amino acids in proteins has been largely elucidated by the use of synthetic polyri- bonucleotides.2-7 For these triplet nucleotide compositions (Table 1), it is of in- terest to examine some of the presently known cases of amino acid substitutions in polypeptides or proteins of known structure. -

Table 2 Amino Acid Sequence of OC-17 As Taken from Ref. 28 Amino
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics This journal is © The Owner Societies 2012 Table 2 Amino Acid Sequence of OC-17 as taken from ref. 28 Amino Acid Abbr. Sequence Position Amino Acid Abbr. Sequence Position Amino Acid Abbr. Sequence Position Aspartic Acid ASP 1 Alanine ALA 49 Arginine ARG 97 Proline PRO 2 Glutamic Acid GLU 50 Phenyalanine PHE 98 Aspartic Acid ASP 3 Leucine LEU 51 Alanine ALA 99 Glycine GLY 4 Arginine ARG 52 Serine SER 100 Cysteine CYS 5 Leucine LEU 53 Tryptophan TRP 101 Glycine GLY 6 Leucine LEU 54 Histidine HIE 102 Proline PRO 7 Alanine ALA 55 Arginine ARG 103 Glycine GLY 8 Glutamic Acid GLU 56 Threonine THR 104 Tryptophan TRP 9 Leucine LEU 57 Alanine ALA 105 Valine VAL 10 Leucine LEU 58 Lysine LYS 106 Proline PRO 11 Asparagine ASN 59 Alanine ALA 107 Threonine THR 12 Alanine ALA 60 Arginine ARG 108 Proline PRO 13 Serine SER 61 Arginine ARG 109 Glycine GLY 14 Arginine ARG 62 Glycine GLY 110 Glycine GLY 15 Glycine GLY 63 Glycine GLY 111 Cysteine CYS 16 Glycine GLY 64 Arginine ARG 112 Leucine LEU 17 Aspartic Acid ASP 65 Cysteine CYS 113 Glycine GLY 18 Glycine GLY 66 Alanine ALA 114 Phenyalanine PHE 19 Serine SER 67 Alanine ALA 115 Phenyalanine PHE 20 Glycine GLY 68 Leucine LEU 116 Serine SER 21 Glutamic Acid GLU 69 Arginine ARG 117 Arginine ARG 22 Glycine GLY 70 Aspartic Acid ASP 118 Glutamic Acid GLU 23 Alanine ALA 71 Glutamic Acid GLU 119 Leucine LEU 24 Aspartic Acid ASP 72 Glutamic Acid GLU 120 Serine SER 25 Glycine GLY 73 Alanine ALA 121 Tryptophan TRP 26 Arginine ARG 74 Phenyalanine -

The Role of Reduced Methionine in Mediating the Metabolic Responses to Protein Restriction Using Different Sources of Protein
nutrients Article The Role of Reduced Methionine in Mediating the Metabolic Responses to Protein Restriction Using Different Sources of Protein Han Fang 1 , Kirsten P. Stone 1 , Sujoy Ghosh 2,3 , Laura A. Forney 4 and Thomas W. Gettys 1,* 1 Laboratory of Nutrient Sensing & Adipocyte Signaling, 6400 Perkins Road, Pennington Biomedical Research Center, Baton Rouge, LA 70808, USA; [email protected] (H.F.); [email protected] (K.P.S.) 2 Laboratory of Computational Biology, Pennington Biomedical Research Center, Baton Rouge, LA 70808, USA; [email protected] 3 Program in Cardiovascular and Metabolic Disorders and Center for Computational Biology, Duke-NUS Medical School, Singapore 169857, Singapore 4 Department of Integrative Biology and Pharmacology, University of Texas Health Science Center at Houston, 7000 Fannin St, Houston, TX 77030, USA; [email protected] * Correspondence: [email protected] Abstract: Dietary protein restriction and dietary methionine restriction (MR) produce a comparable series of behavioral, physiological, biochemical, and transcriptional responses. Both dietary regimens produce a similar reduction in intake of sulfur amino acids (e.g., methionine and cystine), and both diets increase expression and release of hepatic FGF21. Given that FGF21 is an essential mediator of the metabolic phenotype produced by both diets, an important unresolved question is whether dietary protein restriction represents de facto methionine restriction. Using diets formulated from either casein or soy protein with matched reductions in sulfur amino acids, we compared the ability Citation: Fang, H.; Stone, K.P.; of the respective diets to recapitulate the metabolic phenotype produced by methionine restriction Ghosh, S.; Forney, L.A.; Gettys, T.W.