M-Cl-09(Bt) 1 P.T.O
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
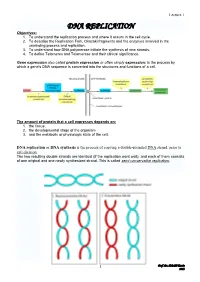
DNA REPLICATION Objectives: 1
Lecture 1 DNA REPLICATION Objectives: 1. To understand the replication process and where it occurs in the cell cycle. 2. To describe the Replication Fork, Okazaki fragments and the enzymes involved in the unwinding process and replication. 3. To understand how DNA polymerase initiate the synthesis of new strands. 4. To define Telomeres and Telomerase and their clinical significance. Gene expression also called protein expression or often simply expression: is the process by which a gene's DNA sequence is converted into the structures and functions of a cell. The amount of protein that a cell expresses depends on: 1. the tissue, 2. the developmental stage of the organism 3. and the metabolic or physiologic state of the cell. DNA replication or DNA synthesis is the process of copying a double-stranded DNA strand, prior to cell division. The two resulting double strands are identical (if the replication went well), and each of them consists of one original and one newly synthesized strand. This is called semi conservative replication. 1 Prof. Dr. H.D.El-Yassin 2013 Lecture 1 The process of replication consists of three steps, initiation, replication and termination. 1. Prokaryotic replication Basic Requirement for DNA Synthesis 1. Substrates: the four deoxy nucleosides triphosphates are needed as substrates for DNA synthesis. Cleavage of the high-energy phosphate bond between the α and β phosphates provides the energy for the addition of the nucleotide. 2. Template: DNA replication cannot occur without a template. A template is required to direct the addition of the appropriate complementary deoxynucleotide to the newly synthesized DNA strand. -
DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli
DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Kath, James Evon. 2016. DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli. Doctoral dissertation, Harvard University, Graduate School of Arts & Sciences. Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:26718716 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA ! ! ! ! ! ! ! DNA!polymerase!exchange!and!lesion!bypass!in!Escherichia)coli! ! A!dissertation!presented! by! James!Evon!Kath! to! The!Committee!on!Higher!Degrees!in!Biophysics! ! in!partial!fulfillment!of!the!requirements! for!the!degree!of! Doctor!of!Philosophy! in!the!subject!of! Biophysics! ! Harvard!University! Cambridge,!Massachusetts! October!2015! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ©!2015!L!James!E.!Kath.!Some!Rights!Reserved.! ! This!work!is!licensed!under!the!Creative!Commons!Attribution!3.0!United!States!License.!To! view!a!copy!of!this!license,!visit:!http://creativecommons.org/licenses/By/3.0/us! ! ! Dissertation!Advisor:!Professor!Joseph!J.!Loparo! ! ! !!!!!!!!James!Evon!Kath! ! DNA$polymerase$exchange$and$lesion$bypass$in$Escherichia)coli$ $ Abstract$ ! Translesion! synthesis! (TLS)! alleviates! -

Phosphatidylinositol-3-Kinase Related Kinases (Pikks) in Radiation-Induced Dna Damage
Mil. Med. Sci. Lett. (Voj. Zdrav. Listy) 2012, vol. 81(4), p. 177-187 ISSN 0372-7025 DOI: 10.31482/mmsl.2012.025 REVIEW ARTICLE PHOSPHATIDYLINOSITOL-3-KINASE RELATED KINASES (PIKKS) IN RADIATION-INDUCED DNA DAMAGE Ales Tichy 1, Kamila Durisova 1, Eva Novotna 1, Lenka Zarybnicka 1, Jirina Vavrova 1, Jaroslav Pejchal 2, Zuzana Sinkorova 1 1 Department of Radiobiology, Faculty of Health Sciences in Hradec Králové, University of Defence in Brno, Czech Republic 2 Centrum of Advanced Studies, Faculty of Health Sciences in Hradec Králové, University of Defence in Brno, Czech Republic. Received 5 th September 2012. Revised 27 th November 2012. Published 7 th December 2012. Summary This review describes a drug target for cancer therapy, family of phosphatidylinositol-3 kinase related kinases (PIKKs), and it gives a comprehensive review of recent information. Besides general information about phosphatidylinositol-3 kinase superfamily, it characterizes a DNA-damage response pathway since it is monitored by PIKKs. Key words: PIKKs; ATM; ATR; DNA-PK; Ionising radiation; DNA-repair ABBREVIATIONS therapy and radiation play a pivotal role. Since cancer is one of the leading causes of death worldwide, it is DSB - double stand breaks, reasonable to invest time and resources in the enligh - IR - ionising radiation, tening of mechanisms, which underlie radio-resis - p53 - TP53 tumour suppressors, tance. PI - phosphatidylinositol. The aim of this review is to describe the family INTRODUCTION of phosphatidyinositol 3-kinases (PI3K) and its func - tional subgroup - phosphatidylinositol-3-kinase rela - An efficient cancer treatment means to restore ted kinases (PIKKs) and their relation to repairing of controlled tissue growth via interfering with cell sig - radiation-induced DNA damage. -

Collision with Duplex DNA Renders Escherichia Coli DNA Polymerase III
www.nature.com/scientificreports OPEN Collision with duplex DNA renders Escherichia coli DNA polymerase III holoenzyme susceptible to Received: 8 May 2017 Accepted: 18 September 2017 DNA polymerase IV-mediated Published: xx xx xxxx polymerase switching on the sliding clamp Thanh Thi Le, Asako Furukohri, Masahiro Tatsumi-Akiyama & Hisaji Maki Organisms possess multiple DNA polymerases (Pols) and use each for a different purpose. One of the five Pols inEscherichia coli, DNA polymerase IV (Pol IV), encoded by the dinB gene, is known to participate in lesion bypass at certain DNA adducts. To understand how cells choose Pols when the replication fork encounters an obstacle on template DNA, the process of polymerase exchange from the primary replicative enzyme DNA polymerase III (Pol III) to Pol IV was studied in vitro. Replicating Pol III forming a tight holoenzyme (Pol III HE) with the sliding clamp was challenged by Pol IV on a primed ssDNA template carrying a short inverted repeat. A rapid and lesion-independent switch from Pol III to Pol IV occurred when Pol III HE encountered a hairpin stem duplex, implying that the loss of Pol III-ssDNA contact induces switching to Pol IV. Supporting this idea, mutant Pol III with an increased affinity for ssDNA was more resistant to Pol IV than wild-type Pol III was. We observed that an exchange between Pol III and Pol IV also occurred when Pol III HE collided with primer/template duplex. Our data suggest that Pol III-ssDNA interaction may modulate the susceptibility of Pol III HE to Pol IV-mediated polymerase exchange. -

Conversion of OX174 and Fd Single-Stranded DNA to Replicative Forms in Extracts of Escherichia Coli (Dnac, Dnad, and Dnag Gene Products/DNA Polymerase III) REED B
Proc. Nat. Acad. Sci. USA Vol. 69, No. 11, pp. 3233-3237, November 1972 Conversion of OX174 and fd Single-Stranded DNA to Replicative Forms in Extracts of Escherichia coli (dnaC, dnaD, and dnaG gene products/DNA polymerase III) REED B. WICKNER, MICHEL WRIGHT, SUE WICKNER, AND JERARD HURWITZ Department of Developmental Biology and Cancer, Division of Biological Sciences, Albert Einstein College of Medicine, Bronx, New York 10461 Communicated by Harry Eagle, August 28, 1972 ABSTRACT 4X174 and M13 (fd) single-stranded cir- MATERIALS AND METHODS cular DNAs are converted to their replicative forms by ex- tracts of E. coli pol Al cells. We find that the qX174 DNA- [a-32P]dTTP was obtained from New England Nuclear Corp. dependent reaction requires Mg++, ATP, and all four de- OX174 DNA was prepared by the method of Sinsheimer (7) oxynucleoside triphosphates, but not CTP, UTP, or GTP. or Franke and Ray (8), while fd viral DNA was prepared as This reaction also involves the products of the dnaC, dnaD, dnaE (DNA polymerase III), and dnaG genes, described (9). Pancreatic RNase was the highest grade ob- but not that of dnaF (ribonucleotide reductase). The in tainable from Worthington Biochemical Corp. It was further vitro conversion of fd single-stranded DNA to the replica- freed of possible contaminating DNase by heating a solution tive form requires all four ribonucleoside triphosphates, (2 mg/ml in 15 mM sodium citrate, pH 5) at 800 for 10 min. Mg++, and all four deoxynucleoside triphosphates. The E. protein was purified from E. coli strain reaction involves the product of gene dnaE but not those coli unwinding of genes dnaC, dnaD, dnaF, or dnaG. -

The Microbiota-Produced N-Formyl Peptide Fmlf Promotes Obesity-Induced Glucose
Page 1 of 230 Diabetes Title: The microbiota-produced N-formyl peptide fMLF promotes obesity-induced glucose intolerance Joshua Wollam1, Matthew Riopel1, Yong-Jiang Xu1,2, Andrew M. F. Johnson1, Jachelle M. Ofrecio1, Wei Ying1, Dalila El Ouarrat1, Luisa S. Chan3, Andrew W. Han3, Nadir A. Mahmood3, Caitlin N. Ryan3, Yun Sok Lee1, Jeramie D. Watrous1,2, Mahendra D. Chordia4, Dongfeng Pan4, Mohit Jain1,2, Jerrold M. Olefsky1 * Affiliations: 1 Division of Endocrinology & Metabolism, Department of Medicine, University of California, San Diego, La Jolla, California, USA. 2 Department of Pharmacology, University of California, San Diego, La Jolla, California, USA. 3 Second Genome, Inc., South San Francisco, California, USA. 4 Department of Radiology and Medical Imaging, University of Virginia, Charlottesville, VA, USA. * Correspondence to: 858-534-2230, [email protected] Word Count: 4749 Figures: 6 Supplemental Figures: 11 Supplemental Tables: 5 1 Diabetes Publish Ahead of Print, published online April 22, 2019 Diabetes Page 2 of 230 ABSTRACT The composition of the gastrointestinal (GI) microbiota and associated metabolites changes dramatically with diet and the development of obesity. Although many correlations have been described, specific mechanistic links between these changes and glucose homeostasis remain to be defined. Here we show that blood and intestinal levels of the microbiota-produced N-formyl peptide, formyl-methionyl-leucyl-phenylalanine (fMLF), are elevated in high fat diet (HFD)- induced obese mice. Genetic or pharmacological inhibition of the N-formyl peptide receptor Fpr1 leads to increased insulin levels and improved glucose tolerance, dependent upon glucagon- like peptide-1 (GLP-1). Obese Fpr1-knockout (Fpr1-KO) mice also display an altered microbiome, exemplifying the dynamic relationship between host metabolism and microbiota. -

Product Sheet Info
Master Clone List for NR-19279 ® Vibrio cholerae Gateway Clone Set, Recombinant in Escherichia coli, Plates 1-46 Catalog No. NR-19279 Table 1: Vibrio cholerae Gateway® Clones, Plate 1 (NR-19679) Clone ID Well ORF Locus ID Symbol Product Accession Position Length Number 174071 A02 367 VC2271 ribD riboflavin-specific deaminase NP_231902.1 174346 A03 336 VC1877 lpxK tetraacyldisaccharide 4`-kinase NP_231511.1 174354 A04 342 VC0953 holA DNA polymerase III, delta subunit NP_230600.1 174115 A05 388 VC2085 sucC succinyl-CoA synthase, beta subunit NP_231717.1 174310 A06 506 VC2400 murC UDP-N-acetylmuramate--alanine ligase NP_232030.1 174523 A07 132 VC0644 rbfA ribosome-binding factor A NP_230293.2 174632 A08 322 VC0681 ribF riboflavin kinase-FMN adenylyltransferase NP_230330.1 174930 A09 433 VC0720 phoR histidine protein kinase PhoR NP_230369.1 174953 A10 206 VC1178 conserved hypothetical protein NP_230823.1 174976 A11 213 VC2358 hypothetical protein NP_231988.1 174898 A12 369 VC0154 trmA tRNA (uracil-5-)-methyltransferase NP_229811.1 174059 B01 73 VC2098 hypothetical protein NP_231730.1 174075 B02 82 VC0561 rpsP ribosomal protein S16 NP_230212.1 174087 B03 378 VC1843 cydB-1 cytochrome d ubiquinol oxidase, subunit II NP_231477.1 174099 B04 383 VC1798 eha eha protein NP_231433.1 174294 B05 494 VC0763 GTP-binding protein NP_230412.1 174311 B06 314 VC2183 prsA ribose-phosphate pyrophosphokinase NP_231814.1 174603 B07 108 VC0675 thyA thymidylate synthase NP_230324.1 174474 B08 466 VC1297 asnS asparaginyl-tRNA synthetase NP_230942.2 174933 B09 198 -

Roles of DNA Polymerases V and II in SOS-Induced Error-Prone and Error-Free Repair in Escherichia Coli
Colloquium Roles of DNA polymerases V and II in SOS-induced error-prone and error-free repair in Escherichia coli Phuong Pham*, Savithri Rangarajan*, Roger Woodgate†, and Myron F. Goodman*‡ *Departments of Biological Sciences and Chemistry, Hedco Molecular Biology Laboratories, University of Southern California, Los Angeles, CA 90089-1340; and †Section on DNA Replication, Repair and Mutagenesis, National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD 20892-2725 DNA polymerase V, composed of a heterotrimer of the DNA regulated at the transcriptional level by the LexA protein (14). damage-inducible UmuC and UmuD2 proteins, working in conjunc- Many of these genes encode proteins required to repair DNA tion with RecA, single-stranded DNA (ssDNA)-binding protein damage (15). The overriding importance of DNA repair is (SSB),  sliding clamp, and ␥ clamp loading complex, are respon- apparent from the observation that a single pyrimidine dimer is sible for most SOS lesion-targeted mutations in Escherichia coli,by lethal in E. coli strains defective for excision and recombinational catalyzing translesion synthesis (TLS). DNA polymerase II, the repair (16). These experiments were among the first to demon- product of the damage-inducible polB (dinA ) gene plays a pivotal strate the essential contribution of DNA repair to cell survival. role in replication-restart, a process that bypasses DNA damage in Excision and recombination repair pathways are referred to as an error-free manner. Replication-restart takes place almost im- ‘‘error-free’’ because they do not result in an increase in muta- mediately after the DNA is damaged (Ϸ2 min post-UV irradiation), tion rate above spontaneous background levels (1). -

Supplementary Information
Supplementary information (a) (b) Figure S1. Resistant (a) and sensitive (b) gene scores plotted against subsystems involved in cell regulation. The small circles represent the individual hits and the large circles represent the mean of each subsystem. Each individual score signifies the mean of 12 trials – three biological and four technical. The p-value was calculated as a two-tailed t-test and significance was determined using the Benjamini-Hochberg procedure; false discovery rate was selected to be 0.1. Plots constructed using Pathway Tools, Omics Dashboard. Figure S2. Connectivity map displaying the predicted functional associations between the silver-resistant gene hits; disconnected gene hits not shown. The thicknesses of the lines indicate the degree of confidence prediction for the given interaction, based on fusion, co-occurrence, experimental and co-expression data. Figure produced using STRING (version 10.5) and a medium confidence score (approximate probability) of 0.4. Figure S3. Connectivity map displaying the predicted functional associations between the silver-sensitive gene hits; disconnected gene hits not shown. The thicknesses of the lines indicate the degree of confidence prediction for the given interaction, based on fusion, co-occurrence, experimental and co-expression data. Figure produced using STRING (version 10.5) and a medium confidence score (approximate probability) of 0.4. Figure S4. Metabolic overview of the pathways in Escherichia coli. The pathways involved in silver-resistance are coloured according to respective normalized score. Each individual score represents the mean of 12 trials – three biological and four technical. Amino acid – upward pointing triangle, carbohydrate – square, proteins – diamond, purines – vertical ellipse, cofactor – downward pointing triangle, tRNA – tee, and other – circle. -

DNA Polymerase I- Dependent Replication (Temperature-Sensitive Dna Mutants/Extragenic Suppression) OSAMI NIWA*, SHARON K
Proc. Nati Acad. Sci. USA Vol. 78, No. 11, pp. 7024-7027, November 1981 Genetics Alternate pathways of DNA replication: DNA polymerase I- dependent replication (temperature-sensitive dna mutants/extragenic suppression) OSAMI NIWA*, SHARON K. BRYAN, AND ROBB E. MOSES Department ofCell Biology, Baylor College of Medicine, Houston, Texas 77030 Communicated by D. Nathans, July 10, 1981 ABSTRACT We have previously shown that someEscherichia proceed in the presence of a functional DNA polymerase I ac- coli [derivatives of strain HS432 (polAl, polB100, polC1026)] can tivity, despite a ts DNA polymerase III (6). replicate DNA at a restrictive temperature in the presence of a We report here that DNA replication in the parent strain polCts mutation and that such revertants contain apparent DNA becomes temperature-resistant with introduction ofDNA poly- polymerase I activity. We demonstrate here that this strain ofE. merase I activity but is ts in the absence of DNA polymerase colibecomes temperature-resistant upon the introduction ofa nor- I or presence of a ts DNA polymerase I activity. We conclude mal gene for DNA polymerase I or suppression of the polAl non- that this strain contains a sense mutation. Such temperature-resistant phenocopies become mutation (pcbA-) that allows repli- temperature-sensitive upon introduction of a temperature-sensi- cation to be dependent on DNA polymerase I polymerizing tive DNA polymerase I gene. Our results confirm that DNA rep- activity. This locus can be transduced to other E. coli strains and lication is DNA polymerase I-dependent in the temperature-re- again exerts phenotypic suppression of the polCts mutation in sistant revertants, indicating that an alternative pathway of the presence of DNA polymerase I. -

Q 297 Suppl USE
The following supplement accompanies the article Atlantic salmon raised with diets low in long-chain polyunsaturated n-3 fatty acids in freshwater have a Mycoplasma dominated gut microbiota at sea Yang Jin, Inga Leena Angell, Simen Rød Sandve, Lars Gustav Snipen, Yngvar Olsen, Knut Rudi* *Corresponding author: [email protected] Aquaculture Environment Interactions 11: 31–39 (2019) Table S1. Composition of high- and low LC-PUFA diets. Stage Fresh water Sea water Feed type High LC-PUFA Low LC-PUFA Fish oil Initial fish weight (g) 0.2 0.4 1 5 15 30 50 0.2 0.4 1 5 15 30 50 80 200 Feed size (mm) 0.6 0.9 1.3 1.7 2.2 2.8 3.5 0.6 0.9 1.3 1.7 2.2 2.8 3.5 3.5 4.9 North Atlantic fishmeal (%) 41 40 40 40 40 30 30 41 40 40 40 40 30 30 35 25 Plant meals (%) 46 45 45 42 40 49 48 46 45 45 42 40 49 48 39 46 Additives (%) 3.3 3.2 3.2 3.5 3.3 3.4 3.9 3.3 3.2 3.2 3.5 3.3 3.4 3.9 2.6 3.3 North Atlantic fish oil (%) 9.9 12 12 15 16 17 18 0 0 0 0 0 1.2 1.2 23 26 Linseed oil (%) 0 0 0 0 0 0 0 6.8 8.1 8.1 9.7 11 10 11 0 0 Palm oil (%) 0 0 0 0 0 0 0 3.2 3.8 3.8 5.4 5.9 5.8 5.9 0 0 Protein (%) 56 55 55 51 49 47 47 56 55 55 51 49 47 47 44 41 Fat (%) 16 18 18 21 22 22 22 16 18 18 21 22 22 22 28 31 EPA+DHA (% diet) 2.2 2.4 2.4 2.9 3.1 3.1 3.1 0.7 0.7 0.7 0.7 0.7 0.7 0.7 4 4.2 Table S2. -

Biotechnology DNA Polymerases
Paper No. : 04 Genetic engineering and recombinant DNA technology Module : 07 DNA polymerases Principal Investigator: Dr Vibha Dhawan, Distinguished Fellow and Sr. Director The Energy and Resouurces Institute (TERI), New Delhi Co-Principal Investigator: Prof S K Jain, Professor, of Medical Biochemistry Jamia Hamdard University, New Delhi Paper Coordinator: Dr Mohan Chandra Joshi, Assistant Professor, Jamia Millia Islamia, New Delhi Content Writer: Dr Samer Singh, Assistant Professor, Panjab University, Chandigarh Content Reviwer: Dr Mohan Chandra Joshi, Assistant Professor, Jamia Millia Islamia, New Delhi Genetic engineering and recombinant DNA technology Biotechnology DNA polymerases Description of Module Subject Name Biotechnology Paper Name Genetic engineering and recombinant DNA technology Module Name/Title DNA polymerases Module Id 07 Pre-requisites Basic DNA Structure and DNA replication Objectives 1) WHAT ARE DNA POLYMERASES? - DISCOVERY 2) TYPES in Escherichia coli 3) WHAT THEY DO? 4) STRUCTURE - CONSERVATION & FUNCTION 5) DNA POLYMERIZATION REACTION and ITS ATTRIBUTES 6) DNA POLYMERASE'S FIDELITY AND PROCESSIVITY 7) DNA POLYMERASE FAMILY AND EXAMPLES 8) HOW SPECIAL FUNCTION DNA POLYMERASE 'TELOMERASE' CARRIES OUT LENGTHENING OF LINEAR DNA ENDS Keywords DNA polymerase, reverse transcriptase, DNA polymerization, Processivity, Fidelity, Telomerase Genetic engineering and recombinant DNA technology Biotechnology DNA polymerases Table of contents A. Learning Objectives B. Keywords C. DNA POLYMERASES C1. General Overview -Discovery C2. Comparison of DNA polymerases of Escherichia coli C3. DNA POLYMERASE CORE STRUCTURE & FUNCTION C4. DNA POLYMERIZATION ACTIVITY and ITS ATTRIBUTES C5. DNA POLYMERASE FAMILIES C6. TELOMERASE- A special function DNA polymerase in eukaryotes D. Summary A. LEARNING OBJECTIVES In this module, Students will learn following: 1. WHAT ARE DNA POLYMERASES? - DISCOVERY 2.