Table 4. V. Cholerae Flexgene ORF Collection
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
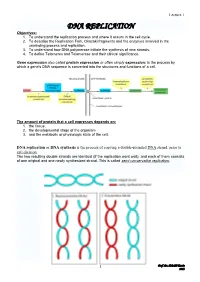
DNA REPLICATION Objectives: 1
Lecture 1 DNA REPLICATION Objectives: 1. To understand the replication process and where it occurs in the cell cycle. 2. To describe the Replication Fork, Okazaki fragments and the enzymes involved in the unwinding process and replication. 3. To understand how DNA polymerase initiate the synthesis of new strands. 4. To define Telomeres and Telomerase and their clinical significance. Gene expression also called protein expression or often simply expression: is the process by which a gene's DNA sequence is converted into the structures and functions of a cell. The amount of protein that a cell expresses depends on: 1. the tissue, 2. the developmental stage of the organism 3. and the metabolic or physiologic state of the cell. DNA replication or DNA synthesis is the process of copying a double-stranded DNA strand, prior to cell division. The two resulting double strands are identical (if the replication went well), and each of them consists of one original and one newly synthesized strand. This is called semi conservative replication. 1 Prof. Dr. H.D.El-Yassin 2013 Lecture 1 The process of replication consists of three steps, initiation, replication and termination. 1. Prokaryotic replication Basic Requirement for DNA Synthesis 1. Substrates: the four deoxy nucleosides triphosphates are needed as substrates for DNA synthesis. Cleavage of the high-energy phosphate bond between the α and β phosphates provides the energy for the addition of the nucleotide. 2. Template: DNA replication cannot occur without a template. A template is required to direct the addition of the appropriate complementary deoxynucleotide to the newly synthesized DNA strand. -
DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli
DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Kath, James Evon. 2016. DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli. Doctoral dissertation, Harvard University, Graduate School of Arts & Sciences. Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:26718716 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA ! ! ! ! ! ! ! DNA!polymerase!exchange!and!lesion!bypass!in!Escherichia)coli! ! A!dissertation!presented! by! James!Evon!Kath! to! The!Committee!on!Higher!Degrees!in!Biophysics! ! in!partial!fulfillment!of!the!requirements! for!the!degree!of! Doctor!of!Philosophy! in!the!subject!of! Biophysics! ! Harvard!University! Cambridge,!Massachusetts! October!2015! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ©!2015!L!James!E.!Kath.!Some!Rights!Reserved.! ! This!work!is!licensed!under!the!Creative!Commons!Attribution!3.0!United!States!License.!To! view!a!copy!of!this!license,!visit:!http://creativecommons.org/licenses/By/3.0/us! ! ! Dissertation!Advisor:!Professor!Joseph!J.!Loparo! ! ! !!!!!!!!James!Evon!Kath! ! DNA$polymerase$exchange$and$lesion$bypass$in$Escherichia)coli$ $ Abstract$ ! Translesion! synthesis! (TLS)! alleviates! -

Folic Acid Antagonists: Antimicrobial and Immunomodulating Mechanisms and Applications
International Journal of Molecular Sciences Review Folic Acid Antagonists: Antimicrobial and Immunomodulating Mechanisms and Applications Daniel Fernández-Villa 1, Maria Rosa Aguilar 1,2 and Luis Rojo 1,2,* 1 Instituto de Ciencia y Tecnología de Polímeros, Consejo Superior de Investigaciones Científicas, CSIC, 28006 Madrid, Spain; [email protected] (D.F.-V.); [email protected] (M.R.A.) 2 Consorcio Centro de Investigación Biomédica en Red de Bioingeniería, Biomateriales y Nanomedicina, 28029 Madrid, Spain * Correspondence: [email protected]; Tel.: +34-915-622-900 Received: 18 September 2019; Accepted: 7 October 2019; Published: 9 October 2019 Abstract: Bacterial, protozoan and other microbial infections share an accelerated metabolic rate. In order to ensure a proper functioning of cell replication and proteins and nucleic acids synthesis processes, folate metabolism rate is also increased in these cases. For this reason, folic acid antagonists have been used since their discovery to treat different kinds of microbial infections, taking advantage of this metabolic difference when compared with human cells. However, resistances to these compounds have emerged since then and only combined therapies are currently used in clinic. In addition, some of these compounds have been found to have an immunomodulatory behavior that allows clinicians using them as anti-inflammatory or immunosuppressive drugs. Therefore, the aim of this review is to provide an updated state-of-the-art on the use of antifolates as antibacterial and immunomodulating agents in the clinical setting, as well as to present their action mechanisms and currently investigated biomedical applications. Keywords: folic acid antagonists; antifolates; antibiotics; antibacterials; immunomodulation; sulfonamides; antimalarial 1. -

(12) Patent Application Publication (10) Pub. No.: US 2013/0089535 A1 Yamashiro Et Al
US 2013 0089535A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2013/0089535 A1 Yamashiro et al. (43) Pub. Date: Apr. 11, 2013 (54) AGENT FOR REDUCING ACETALDEHYDE Publication Classification NORAL CAVITY (51) Int. Cl. (75) Inventors: Kan Yamashiro, Kakamigahara-shi (JP); A68/66 (2006.01) Takahumi Koyama, Kakamigahara-shi A638/51 (2006.01) (JP) A61O 11/00 (2006.01) A638/44 (2006.01) Assignee: AMANOENZYME INC., Nagoya-shi (52) U.S. Cl. (73) CPC. A61K 8/66 (2013.01); A61K 38/44 (2013.01); (JP) A61 K38/51 (2013.01); A61O II/00 (2013.01) (21) Appl. No.: 13/703,451 USPC .......... 424/94.4; 424/94.5; 435/191: 435/232 (22) PCT Fled: Jun. 7, 2011 (57) ABSTRACT Disclosed herein is a novel enzymatic agent effective in (86) PCT NO.: PCT/UP2011/062991 reducing acetaldehyde in the oral cavity. It has been found S371 (c)(1), that an aldehyde dehydrogenase derived from a microorgan (2), (4) Date: Dec. 11, 2012 ism belonging to the genus Saccharomyces and a threonine aldolase derived from Escherichia coli are effective in reduc (30) Foreign Application Priority Data ing low concentrations of acetaldehyde. Therefore, an agent for reducing acetaldehyde in the oral cavity is provided, Jun. 19, 2010 (JP) ................................. 2010-140O26 which contains these enzymes as active ingredients. Patent Application Publication Apr. 11, 2013 Sheet 1 of 2 US 2013/0089535 A1 FIG 1) 10.5 1 0 9.9.5 8. 5 CONTROL TA AD (BSA) ENZYME Patent Application Publication Apr. 11, 2013 Sheet 2 of 2 US 2013/0089535 A1 FIG 2) 110 the CONTROL (BSA) 100 354. -

Enzymatic Encoding Methods for Efficient Synthesis Of
(19) TZZ__T (11) EP 1 957 644 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.: of the grant of the patent: C12N 15/10 (2006.01) C12Q 1/68 (2006.01) 01.12.2010 Bulletin 2010/48 C40B 40/06 (2006.01) C40B 50/06 (2006.01) (21) Application number: 06818144.5 (86) International application number: PCT/DK2006/000685 (22) Date of filing: 01.12.2006 (87) International publication number: WO 2007/062664 (07.06.2007 Gazette 2007/23) (54) ENZYMATIC ENCODING METHODS FOR EFFICIENT SYNTHESIS OF LARGE LIBRARIES ENZYMVERMITTELNDE KODIERUNGSMETHODEN FÜR EINE EFFIZIENTE SYNTHESE VON GROSSEN BIBLIOTHEKEN PROCEDES DE CODAGE ENZYMATIQUE DESTINES A LA SYNTHESE EFFICACE DE BIBLIOTHEQUES IMPORTANTES (84) Designated Contracting States: • GOLDBECH, Anne AT BE BG CH CY CZ DE DK EE ES FI FR GB GR DK-2200 Copenhagen N (DK) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • DE LEON, Daen SK TR DK-2300 Copenhagen S (DK) Designated Extension States: • KALDOR, Ditte Kievsmose AL BA HR MK RS DK-2880 Bagsvaerd (DK) • SLØK, Frank Abilgaard (30) Priority: 01.12.2005 DK 200501704 DK-3450 Allerød (DK) 02.12.2005 US 741490 P • HUSEMOEN, Birgitte Nystrup DK-2500 Valby (DK) (43) Date of publication of application: • DOLBERG, Johannes 20.08.2008 Bulletin 2008/34 DK-1674 Copenhagen V (DK) • JENSEN, Kim Birkebæk (73) Proprietor: Nuevolution A/S DK-2610 Rødovre (DK) 2100 Copenhagen 0 (DK) • PETERSEN, Lene DK-2100 Copenhagen Ø (DK) (72) Inventors: • NØRREGAARD-MADSEN, Mads • FRANCH, Thomas DK-3460 Birkerød (DK) DK-3070 Snekkersten (DK) • GODSKESEN, -

Restriction Endonucleases
Molecular Biology Problem Solver: A Laboratory Guide. Edited by Alan S. Gerstein Copyright © 2001 by Wiley-Liss, Inc. ISBNs: 0-471-37972-7 (Paper); 0-471-22390-5 (Electronic) 9 Restriction Endonucleases Derek Robinson, Paul R. Walsh, and Joseph A. Bonventre Background Information . 226 Which Restriction Enzymes Are Commercially Available? . 226 Why Are Some Enzymes More Expensive Than Others? . 227 What Can You Do to Reduce the Cost of Working with Restriction Enzymes? . 228 If You Could Select among Several Restriction Enzymes for Your Application, What Criteria Should You Consider to Make the Most Appropriate Choice? . 229 What Are the General Properties of Restriction Endonucleases? . 232 What Insight Is Provided by a Restriction Enzyme’s Quality Control Data? . 233 How Stable Are Restriction Enzymes? . 236 How Stable Are Diluted Restriction Enzymes? . 236 Simple Digests . 236 How Should You Set up a Simple Restriction Digest? . 236 Is It Wise to Modify the Suggested Reaction Conditions? . 237 Complex Restriction Digestions . 239 How Can a Substrate Affect the Restriction Digest? . 239 Should You Alter the Reaction Volume and DNA Concentration? . 241 Double Digests: Simultaneous or Sequential? . 242 225 Genomic Digests . 244 When Preparing Genomic DNA for Southern Blotting, How Can You Determine If Complete Digestion Has Been Obtained? . 244 What Are Your Options If You Must Create Additional Rare or Unique Restriction Sites? . 247 Troubleshooting . 255 What Can Cause a Simple Restriction Digest to Fail? . 255 The Volume of Enzyme in the Vial Appears Very Low. Did Leakage Occur during Shipment? . 259 The Enzyme Shipment Sat on the Shipping Dock for Two Days. -

Molecular Markers of Serine Protease Evolution
The EMBO Journal Vol. 20 No. 12 pp. 3036±3045, 2001 Molecular markers of serine protease evolution Maxwell M.Krem and Enrico Di Cera1 ment and specialization of the catalytic architecture should correspond to signi®cant evolutionary transitions in the Department of Biochemistry and Molecular Biophysics, Washington University School of Medicine, Box 8231, St Louis, history of protease clans. Evolutionary markers encoun- MO 63110-1093, USA tered in the sequences contributing to the catalytic apparatus would thus give an account of the history of 1Corresponding author e-mail: [email protected] an enzyme family or clan and provide for comparative analysis with other families and clans. Therefore, the use The evolutionary history of serine proteases can be of sequence markers associated with active site structure accounted for by highly conserved amino acids that generates a model for protease evolution with broad form crucial structural and chemical elements of applicability and potential for extension to other classes of the catalytic apparatus. These residues display non- enzymes. random dichotomies in either amino acid choice or The ®rst report of a sequence marker associated with serine codon usage and serve as discrete markers for active site chemistry was the observation that both AGY tracking changes in the active site environment and and TCN codons were used to encode active site serines in supporting structures. These markers categorize a variety of enzyme families (Brenner, 1988). Since serine proteases of the chymotrypsin-like, subtilisin- AGY®TCN interconversion is an uncommon event, it like and a/b-hydrolase fold clans according to phylo- was reasoned that enzymes within the same family genetic lineages, and indicate the relative ages and utilizing different active site codons belonged to different order of appearance of those lineages. -

Polymerase Ribozyme with Promoter Recognition
In vitro Evolution of a Processive Clamping RNA Polymerase Ribozyme with Promoter Recognition by Razvan Cojocaru BSc, Simon Fraser University, 2014 Thesis Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy in the Department of Molecular Biology and Biochemistry Faculty of Science © Razvan Cojocaru 2021 SIMON FRASER UNIVERSITY Summer 2021 Copyright in this work is held by the author. Please ensure that any reproduction or re-use is done in accordance with the relevant national copyright legislation. Declaration of Committee Name: Razvan Cojocaru Degree: Doctor of Philosophy Title: In vitro Evolution of a Processive Clamping RNA Polymerase Ribozyme with Promoter Recognition Committee: Chair: Lisa Craig Professor, Molecular Biology and Biochemistry Peter Unrau Supervisor Professor, Molecular Biology and Biochemistry Dipankar Sen Committee Member Professor, Molecular Biology and Biochemistry Michel Leroux Committee Member Professor, Molecular Biology and Biochemistry Mani Larijani Internal Examiner Associate Professor, Molecular Biology and Biochemistry Gerald Joyce External Examiner Professor, Jack H. Skirball Center for Chemical Biology and Proteomics Salk Institute for Biological Studies Date Defended/Approved: August 12, 2021 ii Abstract The RNA World hypothesis proposes that the early evolution of life began with RNAs that can serve both as carriers of genetic information and as catalysts. Later in evolution, these functions were gradually replaced by DNA and enzymatic proteins in cellular biology. I start by reviewing the naturally occurring catalytic RNAs, ribozymes, as they play many important roles in biology today. These ribozymes are central to protein synthesis and the regulation of gene expression, creating a landscape that strongly supports an early RNA World. -

Health Effects Support Document for Perfluorooctanoic Acid (PFOA)
United States Office of Water EPA 822-R-16-003 Environmental Protection Mail Code 4304T May 2016 Agency Health Effects Support Document for Perfluorooctanoic Acid (PFOA) Perfluorooctanoic Acid – May 2016 i Health Effects Support Document for Perfluorooctanoic Acid (PFOA) U.S. Environmental Protection Agency Office of Water (4304T) Health and Ecological Criteria Division Washington, DC 20460 EPA Document Number: 822-R-16-003 May 2016 Perfluorooctanoic Acid – May 2016 ii BACKGROUND The Safe Drinking Water Act (SDWA), as amended in 1996, requires the Administrator of the U.S. Environmental Protection Agency (EPA) to periodically publish a list of unregulated chemical contaminants known or anticipated to occur in public water systems and that may require regulation under SDWA. The SDWA also requires the Agency to make regulatory determinations on at least five contaminants on the Contaminant Candidate List (CCL) every 5 years. For each contaminant on the CCL, before EPA makes a regulatory determination, the Agency needs to obtain sufficient data to conduct analyses on the extent to which the contaminant occurs and the risk it poses to populations via drinking water. Ultimately, this information will assist the Agency in determining the most appropriate course of action in relation to the contaminant (e.g., developing a regulation to control it in drinking water, developing guidance, or deciding not to regulate it). The PFOA health assessment was initiated by the Office of Water, Office of Science and Technology in 2009. The draft Health Effects Support Document for Perfluoroctanoic Acid (PFOA) was completed in 2013 and released for public comment in February 2014. -

Serine Proteases with Altered Sensitivity to Activity-Modulating
(19) & (11) EP 2 045 321 A2 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 08.04.2009 Bulletin 2009/15 C12N 9/00 (2006.01) C12N 15/00 (2006.01) C12Q 1/37 (2006.01) (21) Application number: 09150549.5 (22) Date of filing: 26.05.2006 (84) Designated Contracting States: • Haupts, Ulrich AT BE BG CH CY CZ DE DK EE ES FI FR GB GR 51519 Odenthal (DE) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • Coco, Wayne SK TR 50737 Köln (DE) •Tebbe, Jan (30) Priority: 27.05.2005 EP 05104543 50733 Köln (DE) • Votsmeier, Christian (62) Document number(s) of the earlier application(s) in 50259 Pulheim (DE) accordance with Art. 76 EPC: • Scheidig, Andreas 06763303.2 / 1 883 696 50823 Köln (DE) (71) Applicant: Direvo Biotech AG (74) Representative: von Kreisler Selting Werner 50829 Köln (DE) Patentanwälte P.O. Box 10 22 41 (72) Inventors: 50462 Köln (DE) • Koltermann, André 82057 Icking (DE) Remarks: • Kettling, Ulrich This application was filed on 14-01-2009 as a 81477 München (DE) divisional application to the application mentioned under INID code 62. (54) Serine proteases with altered sensitivity to activity-modulating substances (57) The present invention provides variants of ser- screening of the library in the presence of one or several ine proteases of the S1 class with altered sensitivity to activity-modulating substances, selection of variants with one or more activity-modulating substances. A method altered sensitivity to one or several activity-modulating for the generation of such proteases is disclosed, com- substances and isolation of those polynucleotide se- prising the provision of a protease library encoding poly- quences that encode for the selected variants. -

SUPPLEMENTARY INFORMATION in Silico Signature Prediction
SUPPLEMENTARY INFORMATION In Silico Signature Prediction Modeling in Cytolethal Distending Toxin-Producing Escherichia coli Strains Maryam Javadi, Mana Oloomi*, Saeid Bouzari Department of Molecular Biology, Pasteur Institute of Iran, Tehran 13164, Iran http://www.genominfo.org/src/sm/gni-15-69-s001.pdf Supplementary Table 6. Aalphabetic abbreviation and description of putative conserved domains Alphabetic Abbreviation Description 17 Large terminase protein 2_A_01_02 Multidrug resistance protein 2A0115 Benzoate transport; [Transport and binding proteins, Carbohydrates, organic alcohols] 52 DNA topisomerase II medium subunit; Provisional AAA_13 AAA domain; This family of domains contain a P-loop motif AAA_15 AAA ATPase domain; This family of domains contain a P-loop motif AAA_21 AAA domain AAA_23 AAA domain ABC_RecF ATP-binding cassette domain of RecF; RecF is a recombinational DNA repair ATPase ABC_SMC_barmotin ATP-binding cassette domain of barmotin, a member of the SMC protein family AcCoA-C-Actrans Acetyl-CoA acetyltransferases AHBA_syn 3-Amino-5-hydroxybenzoic acid synthase family (AHBA_syn) AidA Type V secretory pathway, adhesin AidA [Cell envelope biogenesis] Ail_Lom Enterobacterial Ail/Lom protein; This family consists of several bacterial and phage Ail_Lom proteins AIP3 Actin interacting protein 3; Aip3p/Bud6p is a regulator of cell and cytoskeletal polarity Aldose_epim_Ec_YphB Aldose 1-epimerase, similar to Escherichia coli YphB AlpA Predicted transcriptional regulator [Transcription] AntA AntA/AntB antirepressor AraC AraC-type -

Supplementary File 2A Revised
Supplementary file 2A. Differentially expressed genes in aldosteronomas compared to all other samples, ranked according to statistical significance. Missing values were not allowed in aldosteronomas, but to a maximum of five in the other samples. Acc UGCluster Name Symbol log Fold Change P - Value Adj. P-Value B R99527 Hs.8162 Hypothetical protein MGC39372 MGC39372 2,17 6,3E-09 5,1E-05 10,2 AA398335 Hs.10414 Kelch domain containing 8A KLHDC8A 2,26 1,2E-08 5,1E-05 9,56 AA441933 Hs.519075 Leiomodin 1 (smooth muscle) LMOD1 2,33 1,3E-08 5,1E-05 9,54 AA630120 Hs.78781 Vascular endothelial growth factor B VEGFB 1,24 1,1E-07 2,9E-04 7,59 R07846 Data not found 3,71 1,2E-07 2,9E-04 7,49 W92795 Hs.434386 Hypothetical protein LOC201229 LOC201229 1,55 2,0E-07 4,0E-04 7,03 AA454564 Hs.323396 Family with sequence similarity 54, member B FAM54B 1,25 3,0E-07 5,2E-04 6,65 AA775249 Hs.513633 G protein-coupled receptor 56 GPR56 -1,63 4,3E-07 6,4E-04 6,33 AA012822 Hs.713814 Oxysterol bining protein OSBP 1,35 5,3E-07 7,1E-04 6,14 R45592 Hs.655271 Regulating synaptic membrane exocytosis 2 RIMS2 2,51 5,9E-07 7,1E-04 6,04 AA282936 Hs.240 M-phase phosphoprotein 1 MPHOSPH -1,40 8,1E-07 8,9E-04 5,74 N34945 Hs.234898 Acetyl-Coenzyme A carboxylase beta ACACB 0,87 9,7E-07 9,8E-04 5,58 R07322 Hs.464137 Acyl-Coenzyme A oxidase 1, palmitoyl ACOX1 0,82 1,3E-06 1,2E-03 5,35 R77144 Hs.488835 Transmembrane protein 120A TMEM120A 1,55 1,7E-06 1,4E-03 5,07 H68542 Hs.420009 Transcribed locus 1,07 1,7E-06 1,4E-03 5,06 AA410184 Hs.696454 PBX/knotted 1 homeobox 2 PKNOX2 1,78 2,0E-06