DNA Polymerase I- Dependent Replication (Temperature-Sensitive Dna Mutants/Extragenic Suppression) OSAMI NIWA*, SHARON K
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Copyright by Young-Sam Lee 2010
Copyright by Young-Sam Lee 2010 The Dissertation Committee for Young-Sam Lee Certifies that this is the approved version of the following dissertation: Structural and Functional Studies of the Human Mitochondrial DNA Polymerase Committee: Whitney Yin, Supervisor Ian Molineux Kenneth Johnson Tanya Paull Jon Robertus Structural and Functional Studies of the Human Mitochondrial DNA Polymerase by Young-Sam Lee, B.S, M.S. Dissertation Presented to the Faculty of the Graduate School of The University of Texas at Austin in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy The University of Texas at Austin August, 2010 Dedication For my wife, In-Sook Jung. Acknowledgements I would like to appreciate Dr. Whitney Yin for giving me chance to working in her lab and mentoring me through my graduate program. Not only the scientific insights, also the warmness that she gave me and my family encouraged me to pursue my Ph. D. degree in the foreign country. I also would like to thank “a guru of molecular biology” Dr. Ian Molineux and “a guru of enzyme kinetics” Dr. Kenneth Johnson. Without their critical advice, I would not be accomplished my publication. I hope to be a respectable expert in my research field like them. I also should remember friendship and generosity given by many current and former Yin lab members: Hey-Ryung Chang, Qingchao “Eric” Meng, Xu Yang, Jeff Knight, Dr. Michio Matsunaga, Dr. He “River” Quan, Taewung Lee, Xin “Ella” Wang, Jamila Momand, and Max Shay. Most of all, I really appreciate my parents for their endless love and support, and my wife, In-Sook Jung, and my son, Jason Seung-Hyeon Lee who always stand by me with patients during my graduate carrier. -
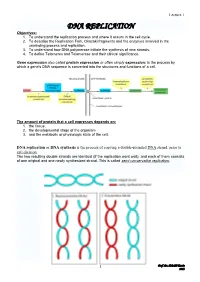
DNA REPLICATION Objectives: 1
Lecture 1 DNA REPLICATION Objectives: 1. To understand the replication process and where it occurs in the cell cycle. 2. To describe the Replication Fork, Okazaki fragments and the enzymes involved in the unwinding process and replication. 3. To understand how DNA polymerase initiate the synthesis of new strands. 4. To define Telomeres and Telomerase and their clinical significance. Gene expression also called protein expression or often simply expression: is the process by which a gene's DNA sequence is converted into the structures and functions of a cell. The amount of protein that a cell expresses depends on: 1. the tissue, 2. the developmental stage of the organism 3. and the metabolic or physiologic state of the cell. DNA replication or DNA synthesis is the process of copying a double-stranded DNA strand, prior to cell division. The two resulting double strands are identical (if the replication went well), and each of them consists of one original and one newly synthesized strand. This is called semi conservative replication. 1 Prof. Dr. H.D.El-Yassin 2013 Lecture 1 The process of replication consists of three steps, initiation, replication and termination. 1. Prokaryotic replication Basic Requirement for DNA Synthesis 1. Substrates: the four deoxy nucleosides triphosphates are needed as substrates for DNA synthesis. Cleavage of the high-energy phosphate bond between the α and β phosphates provides the energy for the addition of the nucleotide. 2. Template: DNA replication cannot occur without a template. A template is required to direct the addition of the appropriate complementary deoxynucleotide to the newly synthesized DNA strand. -
DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli
DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Kath, James Evon. 2016. DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli. Doctoral dissertation, Harvard University, Graduate School of Arts & Sciences. Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:26718716 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA ! ! ! ! ! ! ! DNA!polymerase!exchange!and!lesion!bypass!in!Escherichia)coli! ! A!dissertation!presented! by! James!Evon!Kath! to! The!Committee!on!Higher!Degrees!in!Biophysics! ! in!partial!fulfillment!of!the!requirements! for!the!degree!of! Doctor!of!Philosophy! in!the!subject!of! Biophysics! ! Harvard!University! Cambridge,!Massachusetts! October!2015! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ©!2015!L!James!E.!Kath.!Some!Rights!Reserved.! ! This!work!is!licensed!under!the!Creative!Commons!Attribution!3.0!United!States!License.!To! view!a!copy!of!this!license,!visit:!http://creativecommons.org/licenses/By/3.0/us! ! ! Dissertation!Advisor:!Professor!Joseph!J.!Loparo! ! ! !!!!!!!!James!Evon!Kath! ! DNA$polymerase$exchange$and$lesion$bypass$in$Escherichia)coli$ $ Abstract$ ! Translesion! synthesis! (TLS)! alleviates! -

A Mutation in DNA Polymerase Α Rescues WEE1KO Sensitivity to HU
International Journal of Molecular Sciences Article A Mutation in DNA Polymerase α Rescues WEE1KO Sensitivity to HU Thomas Eekhout 1,2 , José Antonio Pedroza-Garcia 1,2 , Pooneh Kalhorzadeh 1,2, Geert De Jaeger 1,2 and Lieven De Veylder 1,2,* 1 Department of Plant Biotechnology and Bioinformatics, Ghent University, 9052 Gent, Belgium; [email protected] (T.E.); [email protected] (J.A.P.-G.); [email protected] (P.K.); [email protected] (G.D.J.) 2 Center for Plant Systems Biology, VIB, 9052 Gent, Belgium * Correspondence: [email protected] Abstract: During DNA replication, the WEE1 kinase is responsible for safeguarding genomic integrity by phosphorylating and thus inhibiting cyclin-dependent kinases (CDKs), which are the driving force of the cell cycle. Consequentially, wee1 mutant plants fail to respond properly to problems arising during DNA replication and are hypersensitive to replication stress. Here, we report the identification of the pola-2 mutant, mutated in the catalytic subunit of DNA polymerase α, as a suppressor mutant of wee1. The mutated protein appears to be less stable, causing a loss of interaction with its subunits and resulting in a prolonged S-phase. Keywords: replication stress; DNA damage; cell cycle checkpoint Citation: Eekhout, T.; Pedroza- 1. Introduction Garcia, J.A.; Kalhorzadeh, P.; De Jaeger, G.; De Veylder, L. A Mutation DNA replication is a highly complex process that ensures the chromosomes are in DNA Polymerase α Rescues correctly replicated to be passed onto the daughter cells during mitosis. Replication starts WEE1KO Sensitivity to HU. Int. -

PURIFIED THERMOSTABLE NUCLEIC ACID POLYMERASE ENZYME from $I(TERMOTOGA MARITIMA)
Europäisches Patentamt *EP000544789B1* (19) European Patent Office Office européen des brevets (11) EP 0 544 789 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.7: C12N 15/54, C12N 9/12 of the grant of the patent: 05.03.2003 Bulletin 2003/10 (86) International application number: PCT/US91/05753 (21) Application number: 91915802.2 (87) International publication number: (22) Date of filing: 13.08.1991 WO 92/003556 (05.03.1992 Gazette 1992/06) (54) PURIFIED THERMOSTABLE NUCLEIC ACID POLYMERASE ENZYME FROM $i(TERMOTOGA MARITIMA) GEREINIGTES THERMOSTABILES NUKLEINSÄURE-POLYMERASEENZYM AUS THERMOTOGA MARITIMA ENZYME D’ACIDE NUCLEIQUE THERMOSTABLE PURIFIEE PROVENANT DE L’EUBACTERIE $i(THERMOTOGA MARITIMA) (84) Designated Contracting States: (74) Representative: Poredda, Andreas et al AT BE CH DE DK ES FR GB GR IT LI LU NL SE Roche Diagnostics GmbH, Patentabteilung, (30) Priority: 13.08.1990 US 567244 Sandhofer Strasse 116 68305 Mannheim (DE) (43) Date of publication of application: 09.06.1993 Bulletin 1993/23 (56) References cited: • CHEMICAL ABSTRACTS, vol. 105, no. 5, 04 (73) Proprietor: F. HOFFMANN-LA ROCHE AG August 1986, Columbus, OH (US); R. HUBER et 4002 Basel (CH) al., p. 386, AN 38901u • JOURNAL OF BIOLOGICAL CHEMISTRY, vol. (72) Inventors: 264, no. 11, 15 April 1989, American Society for • GELFAND, David, H. Biochemistry & Molecular Biology Inc., Oakland, CA 94611 (US) Baltimore, MD (US); F.C. LAWYER et al., pp. • LAWYER, Frances, C. 6427-6437 Oakland, CA 94611 (US) • STOFFEL, Susanne El Cerrito, CA 94530 (US) Note: Within nine months from the publication of the mention of the grant of the European patent, any person may give notice to the European Patent Office of opposition to the European patent granted. -

Glucokinase Regulatory Protein Is Essential for the Proper Subcellular Localisation of Liver Glucokinase
FEBS Letters 456 (1999) 332^338 FEBS 22420 Glucokinase regulatory protein is essential for the proper subcellular localisation of liver glucokinase Nu¨ria de la Iglesiaa, Maria Veiga-da-Cunhab, Emile Van Schaftingenb, Joan J. Guinovarta, Juan C. Ferrera;* aDepartament de Bioqu|¨mica i Biologia Molecular, Universitat de Barcelona, Mart|¨ i Franque©s, 1, E-08028 Barcelona, Spain bLaboratory of Physiological Chemistry, Christian de Duve Institute of Cellular Pathology and Universite¨ Catholique de Louvain, B-1200 Brussels, Belgium Received in revised form 24 June 1999 pressed by fructose 1-phosphate, both of which bind to Abstract Glucokinase (GK), a key enzyme in the glucose homeostatic responses of the liver, changes its intracellular GKRP and modify its a¤nity for GK [4]. This 68 kDa protein localisation depending on the metabolic status of the cell. Rat is only found in the livers of species that express GK and liver GK and Xenopus laevis GK, fused to the green fluorescent although there is some evidence for its presence in pancreatic protein (GFP), concentrated in the nucleus of cultured rat tissue [5,6], a direct demonstration is still not available. hepatocytes at low glucose and translocated to the cytoplasm at In in vitro assays, rat GKRP can inhibit human pancreatic high glucose. Three mutant forms of Xenopus GK with reduced GK, which shows a high degree of identity to the rat liver affinity for GK regulatory protein (GKRP) did not concentrate isoform [7], as well as Xenopus laevis liver GK [8], a more in the hepatocyte nuclei, even at low glucose. In COS-1 and distantly related protein. -

DNA Polymerase V Activity Is Autoregulated by a Novel Intrinsic DNA-Dependent
1 2 DNA polymerase V activity is autoregulated by a novel intrinsic DNA-dependent 3 ATPase 4 Aysen L. Erdem1, Malgorzata Jaszczur1, Jeffrey G. Bertram1, Roger Woodgate2, Michael M. Cox3 & 5 Myron F. Goodman1 6 1Departments of Biological Sciences and Chemistry, University of Southern California, University 7 Park, Los Angeles, California 90089-2910, USA. 2Laboratory of Genomic Integrity, National 8 Institute of Child Health and Human Development, National Institutes of Health, Bethesda, 9 Maryland 20892-3371, USA. 3Department of Biochemistry, University of Wisconsin-Madison, 10 Madison, Wisconsin 53706, USA. 11 12 Escherichia coli DNA polymerase V (pol V), a heterotrimeric complex composed of UmuD′2C, 13 is marginally active. ATP and RecA play essential roles in the activation of pol V for DNA 14 synthesis including translesion synthesis (TLS). We have established three features of the roles 15 of ATP and RecA. 1) RecA-activated DNA polymerase V (pol V Mut), is a DNA-dependent 16 ATPase; 2) bound ATP is required for DNA synthesis; 3) pol V Mut function is regulated by 17 ATP, with ATP required to bind primer/template (p/t) DNA and ATP hydrolysis triggering 18 dissociation from the DNA. Pol V Mut formed with an ATPase-deficient RecA E38K/K72R 19 mutant hydrolyzes ATP rapidly, establishing the DNA-dependent ATPase as an intrinsic 20 property of pol V Mut distinct from the ATP hydrolytic activity of RecA when bound to 21 single-stranded (ss)DNA as a nucleoprotein filament (RecA*). No similar ATPase activity or 22 autoregulatory mechanism has previously been found for a DNA polymerase. -

Human Glucokinase Gene
Proc. Nati. Acad. Sci. USA Vol. 89, pp. 7698-7702, August 1992 Genetics Human glucokinase gene: Isolation, characterization, and identification of two missense mutations linked to early-onset non-insulin-dependent (type 2) diabetes mellitus (glucose/metabolism/phosphorylation/structure4unctlon/chromosome 7) M. STOFFEL*, PH. FROGUELt, J. TAKEDA*, H. ZOUALItt, N. VIONNET*, S. NISHI*§, I. T. WEBER¶, R. W. HARRISON¶, S. J. PILKISII, S. LESAGEtt, M. VAXILLAIREtt, G. VELHOtt, F. SUNtt, F. lIRSt, PH. PASSAt, D. COHENt, AND G. I. BELL*"** *Howard Hughes Medical Institute, and Departments of Biochemistry and Molecular Biology, and of Medicine, The University of Chicago, 5841 South Maryland Avenue, MC1028, Chicago, IL 60637; §Second Division of Internal Medicine, Hamamatsu University School of Medicine, Hamamatsu, Shizuoka 431-32, Japan; IDepartment of Pharmacology, Jefferson Cancer Institute, Thomas Jefferson University, Philadelphia, PA 19107; IlDepartment of Physiology and Biophysics, State University of New York, Stony Brook, NY 11794; tCentre d'Etude du Polymorphisme Humain, 27 rue Juliette Dodu, and Service d'Endocrinologie, H6pital Saint-Louis, 75010 Paris, France; and tG6ndthon, 1 rue de l'Internationale, 91000 Evry, France Communicated by Jean Dausset, May 28, 1992 ABSTRACT DNA polymorphisms in the glucokinase gene by maintaining a gradient for glucose transport into these cells have recently been shown to be tightly linked to early-onset thereby regulating hepatic glucose disposal. In (3 cells, glu- non-insulin-dependent diabetes mellitus in "80% of French cokinase is believed to be part of the glucose-sensing mech- families with this form of diabetes. We previously identified a anism and to be involved in the regulation ofinsulin secretion. -

Arthur Kornberg Discovered (The First) DNA Polymerase Four
Arthur Kornberg discovered (the first) DNA polymerase Using an “in vitro” system for DNA polymerase activity: 1. Grow E. coli 2. Break open cells 3. Prepare soluble extract 4. Fractionate extract to resolve different proteins from each other; repeat; repeat 5. Search for DNA polymerase activity using an biochemical assay: incorporate radioactive building blocks into DNA chains Four requirements of DNA-templated (DNA-dependent) DNA polymerases • single-stranded template • deoxyribonucleotides with 5’ triphosphate (dNTPs) • magnesium ions • annealed primer with 3’ OH Synthesis ONLY occurs in the 5’-3’ direction Fig 4-1 E. coli DNA polymerase I 5’-3’ polymerase activity Primer has a 3’-OH Incoming dNTP has a 5’ triphosphate Pyrophosphate (PP) is lost when dNMP adds to the chain E. coli DNA polymerase I: 3 separable enzyme activities in 3 protein domains 5’-3’ polymerase + 3’-5’ exonuclease = Klenow fragment N C 5’-3’ exonuclease Fig 4-3 E. coli DNA polymerase I 3’-5’ exonuclease Opposite polarity compared to polymerase: polymerase activity must stop to allow 3’-5’ exonuclease activity No dNTP can be re-made in reversed 3’-5’ direction: dNMP released by hydrolysis of phosphodiester backboneFig 4-4 Proof-reading (editing) of misincorporated 3’ dNMP by the 3’-5’ exonuclease Fidelity is accuracy of template-cognate dNTP selection. It depends on the polymerase active site structure and the balance of competing polymerase and exonuclease activities. A mismatch disfavors extension and favors the exonuclease.Fig 4-5 Superimposed structure of the Klenow fragment of DNA pol I with two different DNAs “Fingers” “Thumb” “Palm” red/orange helix: 3’ in red is elongating blue/cyan helix: 3’ in blue is getting edited Fig 4-6 E. -

Taming the Wild Rubisco: Explorations in Functional Metagenomics
Taming the Wild RubisCO: Explorations in Functional Metagenomics DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Brian Hurin Witte, M.S. Graduate Program in Microbiology The Ohio State University 2012 Dissertation Committee : F. Robert Tabita, Advisor Joseph Krzycki Birgit E. Alber Paul Fuerst Copyright by Brian Hurin Witte 2012 Abstract Ribulose bisphosphate carboxylase/oxygenase (E.C. 4.1.1.39) (RubisCO) is the most abundant protein on Earth and the mechanism by which the vast majority of carbon enters the planet’s biosphere. Despite decades of study, many significant questions about this enzyme remain unanswered. As anthropogenic CO2 levels continue to rise, understanding this key component of the carbon cycle is crucial to forecasting feedback circuits, as well as to engineering food and fuel crops to produce more biomass with few inputs of increasingly scarce resources. This study demonstrates three means of investigating the natural diversity of RubisCO. Chapter 1 builds on existing DNA sequence-based techniques of gene discovery and shows that RubisCO from uncultured organisms can be used to complement growth in a RubisCO-deletion strain of autotrophic bacteria. In a few short steps, the time-consuming work of bringing an autotrophic organism in to pure culture can be circumvented. Chapter 2 details a means of entirely bypassing the bias inherent in sequence-based gene discovery by using selection of RubisCO genes from a metagenomic library. Chapter 3 provides a more in-depth study of the RubisCO from the methanogenic archaeon Methanococcoides burtonii. -

Processivity of DNA Polymerases: Two Mechanisms, One Goal Zvi Kelman1*, Jerard Hurwitz1 and Mike O’Donnell2
Minireview 121 Processivity of DNA polymerases: two mechanisms, one goal Zvi Kelman1*, Jerard Hurwitz1 and Mike O’Donnell2 Replicative DNA polymerases are highly processive Processive DNA synthesis by cellular replicases and the enzymes that polymerize thousands of nucleotides without bacteriophage T4 replicase dissociating from the DNA template. The recently Until recently, the only mechanism for high processivity determined structure of the Escherichia coli bacteriophage that was understood in detail was that utilized by cellular T7 DNA polymerase suggests a unique mechanism that replicases and the replicase of bacteriophage T4. This underlies processivity, and this mechanism may generalize mechanism involves a ring-shaped protein called a ‘DNA to other replicative polymerases. sliding clamp’ that encircles the DNA and tethers the polymerase catalytic unit to the DNA [3,4]. The three- Addresses: 1Department of Molecular Biology, Memorial Sloan- dimensional structures of several sliding clamps have been Kettering Cancer Center, 1275 York Avenue, New York, NY 10021, 2 determined: the eukaryotic proliferating cell nuclear USA and Laboratory of DNA Replication, Howard Hughes Medical β Institute, The Rockefeller University, 1230 York Avenue, New York, NY antigen (PCNA) [5,6]; the subunit of the prokaryotic 10021, USA. DNA polymerase III [7]; and the bacteriophage T4 gene 45 protein (gp45) (J Kuriyan, personal communication) *Corresponding author. (Figure 1). The overall structure of these clamps is very E-mail: [email protected] similar; the PCNA, β subunit and gp45 rings are super- Structure 15 February 1998, 6:121–125 imposable [8]. Each ring has similar dimensions and a http://biomednet.com/elecref/0969212600600121 central cavity large enough to accommodate duplex DNA (Figure 1). -

Enhancing Terminal Deoxynucleotidyl Transferase Activity on Substrates
G C A T T A C G G C A T genes Communication Enhancing Terminal Deoxynucleotidyl Transferase 0 Activity on Substrates with 3 Terminal Structures for Enzymatic De Novo DNA Synthesis 1,2,3, 1,2,3, 1,2,4 Sebastian Barthel y , Sebastian Palluk y, Nathan J. Hillson , 1,2,5,6,7,8,9 1,2,5,10, , Jay D. Keasling and Daniel H. Arlow * y 1 Joint BioEnergy Institute, Emeryville, CA 94608, USA; [email protected] (S.B.); [email protected] (S.P.); [email protected] (N.J.H.); [email protected] (J.D.K.) 2 Biological Systems and Engineering Division, Lawrence Berkeley National Lab, Berkeley, CA 94720, USA 3 Department of Biology, Technische Universität Darmstadt, 64287 Darmstadt, Germany 4 DOE Joint Genome Institute, Walnut Creek, CA 94598, USA 5 Institute for Quantitative Biosciences, UC Berkeley, Berkeley, CA 94720, USA 6 Department of Chemical and Biomolecular Engineering, UC Berkeley, Berkeley, CA 94720, USA 7 Department of Bioengineering UC Berkeley, Berkeley, CA 94720, USA 8 Novo Nordisk Foundation Center for Biosustainability, Technical University of Denmark, 2970 Hørsholm, Denmark 9 Center for Synthetic Biochemistry, Institute for Synthetic Biology, Shenzhen Institutes for Advanced Technologies, Shenzhen 518055, China 10 Biophysics Graduate Group, UC Berkeley, Berkeley, CA 94720, USA * Correspondence: [email protected]; Tel.: +1-248-227-5556 Current address: Ansa Biotechnologies, Berkeley, CA 94170, USA. y Received: 8 December 2019; Accepted: 7 January 2020; Published: 16 January 2020 Abstract: Enzymatic oligonucleotide synthesis methods based on the template-independent polymerase terminal deoxynucleotidyl transferase (TdT) promise to enable the de novo synthesis of long oligonucleotides under mild, aqueous conditions.