Sulfate-Dependent Acetate Oxidation Under Extremely Natron-Alkaline Conditions by Syntrophic Associations from Hypersaline Soda Lakes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Crystalline Iron Oxides Stimulate Methanogenic Benzoate Degradation in Marine Sediment-Derived Enrichment Cultures
The ISME Journal https://doi.org/10.1038/s41396-020-00824-7 ARTICLE Crystalline iron oxides stimulate methanogenic benzoate degradation in marine sediment-derived enrichment cultures 1,2 1 3 1,2 1 David A. Aromokeye ● Oluwatobi E. Oni ● Jan Tebben ● Xiuran Yin ● Tim Richter-Heitmann ● 2,4 1 5 5 1 2,3 Jenny Wendt ● Rolf Nimzyk ● Sten Littmann ● Daniela Tienken ● Ajinkya C. Kulkarni ● Susann Henkel ● 2,4 2,4 1,3 2,3,4 1,2 Kai-Uwe Hinrichs ● Marcus Elvert ● Tilmann Harder ● Sabine Kasten ● Michael W. Friedrich Received: 19 May 2020 / Revised: 9 October 2020 / Accepted: 22 October 2020 © The Author(s) 2020. This article is published with open access Abstract Elevated dissolved iron concentrations in the methanic zone are typical geochemical signatures of rapidly accumulating marine sediments. These sediments are often characterized by co-burial of iron oxides with recalcitrant aromatic organic matter of terrigenous origin. Thus far, iron oxides are predicted to either impede organic matter degradation, aiding its preservation, or identified to enhance organic carbon oxidation via direct electron transfer. Here, we investigated the effect of various iron oxide phases with differing crystallinity (magnetite, hematite, and lepidocrocite) during microbial 1234567890();,: 1234567890();,: degradation of the aromatic model compound benzoate in methanic sediments. In slurry incubations with magnetite or hematite, concurrent iron reduction, and methanogenesis were stimulated during accelerated benzoate degradation with methanogenesis as the dominant electron sink. In contrast, with lepidocrocite, benzoate degradation, and methanogenesis were inhibited. These observations were reproducible in sediment-free enrichments, even after five successive transfers. Genes involved in the complete degradation of benzoate were identified in multiple metagenome assembled genomes. -
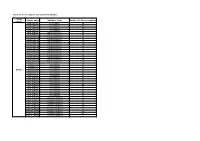
Table S1: List of Samples Included in the Analysis
Table S1: list of samples included in the analysis Study Sample name Inhibitory status Number of days at sampling number DNA.0P2T4 No inhibition 29 DNA.0P2T6 No inhibition 57 DNA.10P2T4 No inhibition 29 DNA.10P2T6 No inhibition 57 DNA.75P2T4 Phenol inhibition 29 DNA.75P2T6 Phenol inhibition 57 DNA.100P2T4 Phenol inhibition 29 DNA.100P2T6 Phenol inhibition 57 DNA.125P1T4 Phenol inhibition 29 DNA.125P1T6 Phenol inhibition 57 DNA.125P2T4 Phenol inhibition 29 DNA.125P2T6 Phenol inhibition 57 DNA.125P3T4 Phenol inhibition 29 DNA.125P3T6 Phenol inhibition 57 DNA.150P2T4 Phenol inhibition 29 DNA.150P2T6 Phenol inhibition 57 DNA.200P2T4 Phenol inhibition 29 DNA.200P2T6 Phenol inhibition 57 DNA.0N2T4 No inhibition 29 DNA.0N2T5 No inhibition 42 DNA.0N2T6 No inhibition 57 Study 1 DNA.5N2T4 No inhibition 29 DNA.5N2T5 No inhibition 42 DNA.5N2T6 No inhibition 57 DNA.10N2T4 No inhibition 29 DNA.10N2T5 No inhibition 42 DNA.10N2T6 No inhibition 57 DNA.15N2T4 No inhibition 29 DNA.15N2T5 No inhibition 42 DNA.15N2T6 No inhibition 57 DNA.25N2T4 No inhibition 29 DNA.25N2T5 No inhibition 42 DNA.25N2T6 No inhibition 57 DNA.75N2T4 Ammonia inhibition 29 DNA.75N2T5 Ammonia inhibition 42 DNA.75N2T6 Ammonia inhibition 57 DNA.100N2T4 Ammonia inhibition 29 DNA.100N2T5 Ammonia inhibition 42 DNA.100N2T6 Ammonia inhibition 57 DNA.250N2T4 Ammonia inhibition 29 DNA.250N2T5 Ammonia inhibition 42 DNA.250N2T6 Ammonia inhibition 57 nono2T3 No inhibition 16 noN2T4 Ammonia inhibition 23 noN2T8 Ammonia inhibition 60 noN2T9 Ammonia inhibition 85 noPhi2T4 Phenol inhibition 23 noPhi2T5 -

Biorremediación De Metales Pesados Por Sulfidogénesis Utilizando Comunidades Y Microorganismos Sulfato-Reductores
FACULTAD DE CIENCIAS EXACTAS DEPARTAMENTO DE QUÍMICA BIORREMEDIACIÓN DE METALES PESADOS POR SULFIDOGÉNESIS UTILIZANDO COMUNIDADES Y MICROORGANISMOS SULFATO-REDUCTORES Trabajo de Tesis Doctoral Graciana Willis Poratti Director: Dr. Edgardo Donati Año 2016 Este Trabajo de Tesis Doctoral es presentado para optar al grado de Doctor de la Facultad de Ciencias Exactas Fue realizado en CINDEFI (CCT La Plata-CONICET, UNLP) A Mamá, Papá y Juan, por acompañarme siempre Reconocimientos A la Facultad de Ciencias Exactas de la Universidad Nacional de La Plata y al Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) por haber hecho posible la realización del presente Trabajo de Tesis Doctoral. Al CINDEFI (Centro de Investigación en Fermentaciones Industriales) y a toda la gente que lo conforma por haberme brindado el lugar físico para la realización del presente trabajo. Al Prof. Dr. David Barrie Johnson (Bangor Acidophile Research Team) de la Universidad de Bangor, Gales por permitirme realizar parte del trabajo de Tesis Doctoral en su laboratorio. A los Dres. Goh Kian Mau de la Universiti Teknologi, Malaysia y Kok-Gan Chan de la University of Malaysa, por la colaboración en la secuenciación del genoma de la cepa LMa1. Al Ente Provincial de Termas de Neuquén (EPROTEN) por permitir la toma de muestra en el Parque Geotermal Copahue. Al grupo de la Dra. Alejandra Giaveno perteneciente al IDEPA (CONICET, UNCo) y a la Facultad de Ingeniería de la Universidad Nacional del Comahue, por la organización de las campañas de muestreo a la zona de Copahue. Agradecimientos A mi director Dr. Edgardo Donati, por haber depositado su confianza en la realización de este trabajo, por dirigirme y haberme dado la oportunidad de viajar y formarme. -

Pila Genes. the Location of Alpha Helices (Represented by Red H) and Beta Strands (Represented by Yellow E) Was Predicted with Jpred 4 (Drozdetskiy Et Al, 2015)
Supplementary Figure S1. Secondary structure of the N-terminus of PilA proteins from Desulfuromondales species and other bacteria that possess type IVa pilA genes. The location of alpha helices (represented by red H) and beta strands (represented by yellow E) was predicted with Jpred 4 (Drozdetskiy et al, 2015). Transmembrane helices (green background) were predicted with TmPred (Hofmann & Stoffel, 1993), TMHMM (Krogh et al, 2001), and HMMTOP (Tusnady & Simon, 2001). G. bemidjiensis (Gbem_2590) MLNKLRSNKGFTLIELLIVVAIIGILAAIAIPQFSAYREKAYNAASNSDLKNFKTGLEAFNADFQTYPAAYVASTN ---HHH----HHHHHHHHHHHHHHHHHHHH----HHHHHHHHHHHHH-----HHHHHHHHHH-------EEEE--- G. bremensis (K419DRAFT_00801) MLNKLRSNKGFTLIELLIVVAIIGILAAIAIPQFSAYREKAYNAASNSDLKNWKTGQEAYQADFQAYPAAYDVH --HHHH----HHHHHHHHHHHHHHHHHHH----HHHHHHHHHH------HHHHHHHHHHHHHHH---------- Pelobacter seleniigenes (N909DRAFT_0006) MLKKFRKNEKGFTLIELLIVVAIIGILAAIAIPQFASYRQKAFNSASQSDLKTIKTSLEGYYTDEYYYPY --HHHHH-----HHHHHHHHHHHHHHHHHHHHH-HHHHHHHHHHHHHHHHHHHHHHHHHHHHH------- Geobacter sp. OR-1 (WP_041974243) MLSKLRSNKGFTLIELLIVVAIIGILAAIAIPQFSAYREKAYNTAANADDKNAKTGEEAYNADNQKYPLAYDQH --HHHHH----HHHHHHHHHHHHHHHHHHH---HHHHHHHHHHHHHHHHHHHHHHHHHHHHH------------ Geobacter sp. M18 (GM18_2492) MLNKIRSNKGFTLIELLIVVAIIGILAAIAIPQFSAYRAKAYNAAANSDLKNIKTGMEAYMADRQAYPVSLDER --HHHHH-----HHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH------------ Geobacter sp. M21 MLNKLRSNKGFTLIELLIVVAIIGILAAIAIPQFSAYRAKAYNSAANSDLKNMKTGMEAYMADRQAYPALLDQR --HHHHH-----HHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH----------- Desulfuromonas -

EXPERIMENTAL STUDIES on FERMENTATIVE FIRMICUTES from ANOXIC ENVIRONMENTS: ISOLATION, EVOLUTION, and THEIR GEOCHEMICAL IMPACTS By
EXPERIMENTAL STUDIES ON FERMENTATIVE FIRMICUTES FROM ANOXIC ENVIRONMENTS: ISOLATION, EVOLUTION, AND THEIR GEOCHEMICAL IMPACTS By JESSICA KEE EUN CHOI A dissertation submitted to the School of Graduate Studies Rutgers, The State University of New Jersey In partial fulfillment of the requirements For the degree of Doctor of Philosophy Graduate Program in Microbial Biology Written under the direction of Nathan Yee And approved by _______________________________________________________ _______________________________________________________ _______________________________________________________ _______________________________________________________ New Brunswick, New Jersey October 2017 ABSTRACT OF THE DISSERTATION Experimental studies on fermentative Firmicutes from anoxic environments: isolation, evolution and their geochemical impacts by JESSICA KEE EUN CHOI Dissertation director: Nathan Yee Fermentative microorganisms from the bacterial phylum Firmicutes are quite ubiquitous in subsurface environments and play an important biogeochemical role. For instance, fermenters have the ability to take complex molecules and break them into simpler compounds that serve as growth substrates for other organisms. The research presented here focuses on two groups of fermentative Firmicutes, one from the genus Clostridium and the other from the class Negativicutes. Clostridium species are well-known fermenters. Laboratory studies done so far have also displayed the capability to reduce Fe(III), yet the mechanism of this activity has not been investigated -

Syntrophic Butyrate and Propionate Oxidation Processes 491
Environmental Microbiology Reports (2010) 2(4), 489–499 doi:10.1111/j.1758-2229.2010.00147.x Minireview Syntrophic butyrate and propionate oxidation processes: from genomes to reaction mechanismsemi4_147 489..499 Nicolai Müller,1† Petra Worm,2† Bernhard Schink,1* a cytoplasmic fumarate reductase to drive energy- Alfons J. M. Stams2 and Caroline M. Plugge2 dependent succinate oxidation. Furthermore, we 1Faculty for Biology, University of Konstanz, D-78457 propose that homologues of the Thermotoga mar- Konstanz, Germany. itima bifurcating [FeFe]-hydrogenase are involved 2Laboratory of Microbiology, Wageningen University, in NADH oxidation by S. wolfei and S. fumaroxidans Dreijenplein 10, 6703 HB Wageningen, the Netherlands. to form hydrogen. Summary Introduction In anoxic environments such as swamps, rice fields In anoxic environments such as swamps, rice paddy fields and sludge digestors, syntrophic microbial communi- and intestines of higher animals, methanogenic commu- ties are important for decomposition of organic nities are important for decomposition of organic matter to matter to CO2 and CH4. The most difficult step is the CO2 and CH4 (Schink and Stams, 2006; Mcinerney et al., fermentative degradation of short-chain fatty acids 2008; Stams and Plugge, 2009). Moreover, they are the such as propionate and butyrate. Conversion of these key biocatalysts in anaerobic bioreactors that are used metabolites to acetate, CO2, formate and hydrogen is worldwide to treat industrial wastewaters and solid endergonic under standard conditions and occurs wastes. Different types of anaerobes have specified only if methanogens keep the concentrations of these metabolic functions in the degradation pathway and intermediate products low. Butyrate and propionate depend on metabolite transfer which is called syntrophy degradation pathways include oxidation steps of (Schink and Stams, 2006). -

The Microbial Sulfur Cycle at Extremely Haloalkaline Conditions of Soda Lakes
REVIEW ARTICLE published: 21 March 2011 doi: 10.3389/fmicb.2011.00044 The microbial sulfur cycle at extremely haloalkaline conditions of soda lakes Dimitry Y. Sorokin1,2*, J. Gijs Kuenen 2 and Gerard Muyzer 2 1 Winogradsky Institute of Microbiology, Russian Academy of Sciences, Moscow, Russia 2 Department of Biotechnology, Delft University of Technology, Delft, Netherlands Edited by: Soda lakes represent a unique ecosystem with extremely high pH (up to 11) and salinity (up to Martin G. Klotz, University of Louisville, saturation) due to the presence of high concentrations of sodium carbonate in brines. Despite USA these double extreme conditions, most of the lakes are highly productive and contain a fully Reviewed by: Aharon Oren, The Hebrew University functional microbial system. The microbial sulfur cycle is among the most active in soda lakes. of Jerusalem, Israel One of the explanations for that is high-energy efficiency of dissimilatory conversions of inorganic Yanhe Ma, Institute of Microbiology sulfur compounds, both oxidative and reductive, sufficient to cope with costly life at double Chinese Academy of Sciences, China extreme conditions. The oxidative part of the sulfur cycle is driven by chemolithoautotrophic *Correspondence: haloalkaliphilic sulfur-oxidizing bacteria (SOB), which are unique for soda lakes. The haloalkaliphilic Dimitry Y. Sorokin, Winogradsky Institute of Microbiology, Russian SOB are present in the surface sediment layer of various soda lakes at high numbers of up to Academy of Sciences, Prospect 60-let 106 viable cells/cm3. The culturable forms are so far represented by four novel genera within the Octyabrya 7/2, 117312 Moscow, Russia Gammaproteobacteria, including the genera Thioalkalivibrio, Thioalkalimicrobium, Thioalkalispira, e-mail: [email protected]; and Thioalkalibacter. -

Bibliography
Bibliography Abella, C.A., X.P. Cristina, A. Martinez, I. Pibernat and X. Vila. 1998. on moderate concentrations of acetate: production of single cells. Two new motile phototrophic consortia: "Chlorochromatium lunatum" Appl. Microbiol. Biotechnol. 35: 686-689. and "Pelochromatium selenoides". Arch. Microbiol. 169: 452-459. Ahring, B.K, P. Westermann and RA. Mah. 1991b. Hydrogen inhibition Abella, C.A and LJ. Garcia-Gil. 1992. Microbial ecology of planktonic of acetate metabolism and kinetics of hydrogen consumption by Me filamentous phototrophic bacteria in holomictic freshwater lakes. Hy thanosarcina thermophila TM-I. Arch. Microbiol. 157: 38-42. drobiologia 243-244: 79-86. Ainsworth, G.C. and P.H.A Sheath. 1962. Microbial Classification: Ap Acca, M., M. Bocchetta, E. Ceccarelli, R Creti, KO. Stetter and P. Cam pendix I. Symp. Soc. Gen. Microbiol. 12: 456-463. marano. 1994. Updating mass and composition of archaeal and bac Alam, M. and D. Oesterhelt. 1984. Morphology, function and isolation terial ribosomes. Archaeal-like features of ribosomes from the deep of halobacterial flagella. ]. Mol. Biol. 176: 459-476. branching bacterium Aquifex pyrophilus. Syst. Appl. Microbiol. 16: 629- Albertano, P. and L. Kovacik. 1994. Is the genus LeptolynglYya (Cyano 637. phyte) a homogeneous taxon? Arch. Hydrobiol. Suppl. 105: 37-51. Achenbach-Richter, L., R Gupta, KO. Stetter and C.R Woese. 1987. Were Aldrich, H.C., D.B. Beimborn and P. Schönheit. 1987. Creation of arti the original eubacteria thermophiles? Syst. Appl. Microbiol. 9: 34- factual internal membranes during fixation of Methanobacterium ther 39. moautotrophicum. Can.]. Microbiol. 33: 844-849. Adams, D.G., D. Ashworth and B. -

'Candidatus Desulfonatronobulbus Propionicus': a First Haloalkaliphilic
Delft University of Technology ‘Candidatus Desulfonatronobulbus propionicus’ a first haloalkaliphilic member of the order Syntrophobacterales from soda lakes Sorokin, D. Y.; Chernyh, N. A. DOI 10.1007/s00792-016-0881-3 Publication date 2016 Document Version Accepted author manuscript Published in Extremophiles: life under extreme conditions Citation (APA) Sorokin, D. Y., & Chernyh, N. A. (2016). ‘Candidatus Desulfonatronobulbus propionicus’: a first haloalkaliphilic member of the order Syntrophobacterales from soda lakes. Extremophiles: life under extreme conditions, 20(6), 895-901. https://doi.org/10.1007/s00792-016-0881-3 Important note To cite this publication, please use the final published version (if applicable). Please check the document version above. Copyright Other than for strictly personal use, it is not permitted to download, forward or distribute the text or part of it, without the consent of the author(s) and/or copyright holder(s), unless the work is under an open content license such as Creative Commons. Takedown policy Please contact us and provide details if you believe this document breaches copyrights. We will remove access to the work immediately and investigate your claim. This work is downloaded from Delft University of Technology. For technical reasons the number of authors shown on this cover page is limited to a maximum of 10. Extremophiles DOI 10.1007/s00792-016-0881-3 ORIGINAL PAPER ‘Candidatus Desulfonatronobulbus propionicus’: a first haloalkaliphilic member of the order Syntrophobacterales from soda lakes D. Y. Sorokin1,2 · N. A. Chernyh1 Received: 23 August 2016 / Accepted: 4 October 2016 © Springer Japan 2016 Abstract Propionate can be directly oxidized anaerobi- from its members at the genus level. -

Sulfate-Reducing Bacteria in Anaerobic Bioreactors Are Presented in Table 1
Sulfate-reducing Bacteria inAnaerobi c Bioreactors Stefanie J.W.H. Oude Elferink Promotoren: dr. ir. G. Lettinga bijzonder hoogleraar ind eanaërobisch e zuiveringstechnologie en hergebruik dr. W.M. deVo s hoogleraar ind e microbiologie Co-promotor: dr. ir. AJ.M. Stams universitair docent bij deleerstoelgroe p microbiologie ^OSJO^-M'3^- Stefanie J.W.H.Oud eElferin k Sulfate-reducing Bacteria inAnaerobi c Bioreactors Proefschrift terverkrijgin g van degraa d van doctor op gezag van derecto r magnificus van deLandbouwuniversitei t Wageningen, dr. C.M. Karssen, inhe t openbaar te verdedigen opvrijda g 22me i 1998 des namiddags tehal f twee ind eAula . r.r, A tri ISBN 90 5485 8451 The research described inthi s thesiswa s financially supported by agran t ofth e Innovative Oriented Program (IOP) Committee on Environmental Biotechnology (IOP-m 90209) established by the Dutch Ministry of Economics, and a grant from Pâques BV. Environmental Technology, P.O. Box 52, 8560AB ,Balk , TheNetherlands . BIBLIOTHEEK LANDBOUWUNIVERSITEIT WAGENTNGEN 1 (J ÜOB^ . ^3"£ Stellingen 1. Inhu n lijst van mogelijke scenario's voor de anaërobe afbraak van propionaat onder sulfaatrijke condities vergeten Uberoi enBhattachary a het scenario dat ind e anaërobe waterzuiveringsreactor van depapierfabrie k teEerbee k lijkt opt etreden , namelijk de afbraak vanpropionaa t door syntrofen en sulfaatreduceerders end e afbraak van acetaat en waterstof door sulfaatreduceerders en methanogenen. Ditproefschrift, hoofdstuk 7 UberoiV, Bhattacharya SK (1995)Interactions among sulfate reducers, acetogens, and methanogens in anaerobicpropionate systems. 2. De stelling van McCartney en Oleszkiewicz dat sulfaatreduceerders inanaërob e reactoren waarschijnlijk alleen competerenme t methanogenen voor het aanwezige waterstof, omdat acetaatafbrekende sulfaatreduceerders nog nooit uit anaëroob slib waren geïsoleerd, was correct bij indiening, maar achterhaald bij publicatie. -

Microbial Communities Mediating Algal Detritus Turnover Under Anaerobic Conditions
Microbial communities mediating algal detritus turnover under anaerobic conditions Jessica M. Morrison1,*, Chelsea L. Murphy1,*, Kristina Baker1, Richard M. Zamor2, Steve J. Nikolai2, Shawn Wilder3, Mostafa S. Elshahed1 and Noha H. Youssef1 1 Department of Microbiology and Molecular Genetics, Oklahoma State University, Stillwater, OK, USA 2 Grand River Dam Authority, Vinita, OK, USA 3 Department of Integrative Biology, Oklahoma State University, Stillwater, OK, USA * These authors contributed equally to this work. ABSTRACT Background. Algae encompass a wide array of photosynthetic organisms that are ubiquitously distributed in aquatic and terrestrial habitats. Algal species often bloom in aquatic ecosystems, providing a significant autochthonous carbon input to the deeper anoxic layers in stratified water bodies. In addition, various algal species have been touted as promising candidates for anaerobic biogas production from biomass. Surprisingly, in spite of its ecological and economic relevance, the microbial community involved in algal detritus turnover under anaerobic conditions remains largely unexplored. Results. Here, we characterized the microbial communities mediating the degradation of Chlorella vulgaris (Chlorophyta), Chara sp. strain IWP1 (Charophyceae), and kelp Ascophyllum nodosum (phylum Phaeophyceae), using sediments from an anaerobic spring (Zodlteone spring, OK; ZDT), sludge from a secondary digester in a local wastewater treatment plant (Stillwater, OK; WWT), and deeper anoxic layers from a seasonally stratified lake -

Quantitative Population Dynamics of Microbial Communities in Plankton-Fed Microbial Fuel Cells
The ISME Journal (2009) 3, 635–646 & 2009 International Society for Microbial Ecology All rights reserved 1751-7362/09 $32.00 www.nature.com/ismej ORIGINAL ARTICLE Quantitative population dynamics of microbial communities in plankton-fed microbial fuel cells Helen K White1, Clare E Reimers2, Erik E Cordes1, Geoffrey F Dilly1 and Peter R Girguis1 1Biological Labs, Department of Organismic and Evolutionary Biology, Harvard University, Cambridge, MA, USA and 2College of Oceanic and Atmospheric Sciences, Hatfield Marine Science Center, Oregon State University, Newport, OR, USA This study examines changes in diversity and abundance of bacteria recovered from the anodes of microbial fuel cells (MFCs) in relation to anode potential, power production and geochemistry. MFCs were batch-fed with plankton, and two systems were maintained at different potentials whereas one was at open circuit for 56.8 days. Bacterial phylogenetic diversity during peak power was assessed from 16S rDNA clone libraries. Throughout the experiment, microbial community structure was examined using terminal restriction fragment length polymorphism. Changes in cell density of key phylotypes, including representatives of d-, e-, c-proteobacteria and Flavobacterium-Cytophaga- Bacteroides, were enumerated by quantitative PCR. Marked differences in phylogenetic diversity were observed during peak power versus the final time point, and changes in microbial community structure were strongly correlated to dissolved organic carbon and ammonium concentrations within the anode chambers. Community structure was notably different between the MFCs at different anode potentials during the onset of peak power. At the final time point, however, the anode-hosted communities in all MFCs were similar. These data demonstrate that differences in growth, succession and population dynamics of key phylotypes were due to anode potential, which may relate to their ability to exploit the anode as an electron acceptor.