Characterization of an Anaerobic Population Digesting a Model Substrate for Maize in the Presence of Trace Metals
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Crystalline Iron Oxides Stimulate Methanogenic Benzoate Degradation in Marine Sediment-Derived Enrichment Cultures
The ISME Journal https://doi.org/10.1038/s41396-020-00824-7 ARTICLE Crystalline iron oxides stimulate methanogenic benzoate degradation in marine sediment-derived enrichment cultures 1,2 1 3 1,2 1 David A. Aromokeye ● Oluwatobi E. Oni ● Jan Tebben ● Xiuran Yin ● Tim Richter-Heitmann ● 2,4 1 5 5 1 2,3 Jenny Wendt ● Rolf Nimzyk ● Sten Littmann ● Daniela Tienken ● Ajinkya C. Kulkarni ● Susann Henkel ● 2,4 2,4 1,3 2,3,4 1,2 Kai-Uwe Hinrichs ● Marcus Elvert ● Tilmann Harder ● Sabine Kasten ● Michael W. Friedrich Received: 19 May 2020 / Revised: 9 October 2020 / Accepted: 22 October 2020 © The Author(s) 2020. This article is published with open access Abstract Elevated dissolved iron concentrations in the methanic zone are typical geochemical signatures of rapidly accumulating marine sediments. These sediments are often characterized by co-burial of iron oxides with recalcitrant aromatic organic matter of terrigenous origin. Thus far, iron oxides are predicted to either impede organic matter degradation, aiding its preservation, or identified to enhance organic carbon oxidation via direct electron transfer. Here, we investigated the effect of various iron oxide phases with differing crystallinity (magnetite, hematite, and lepidocrocite) during microbial 1234567890();,: 1234567890();,: degradation of the aromatic model compound benzoate in methanic sediments. In slurry incubations with magnetite or hematite, concurrent iron reduction, and methanogenesis were stimulated during accelerated benzoate degradation with methanogenesis as the dominant electron sink. In contrast, with lepidocrocite, benzoate degradation, and methanogenesis were inhibited. These observations were reproducible in sediment-free enrichments, even after five successive transfers. Genes involved in the complete degradation of benzoate were identified in multiple metagenome assembled genomes. -

Revised Supplement 1: Reference List for Figure 1
Revised Supplement 1: Reference list for Figure 1. Manuscript title: Process disturbances in agricultural biogas production – causes, mechanisms and effects on the biogas microbiome: A review Susanne Theuerl 1,*, Johanna Klang 1, Annette Prochnow 1,2 1 Leibniz Institute for Agricultural Engineering and Bioeconomy, Max-Exth-Allee 100, 14469 Potsdam, Germany, [email protected] (ST), [email protected] (JK), [email protected] (AP) 2 Humboldt-Universität zu Berlin, Albrecht-Daniel-Thaer-Institute for Agricultural and Horticultural Sciences, Hinter der Reinhardtstr. 6-8, 10115 Berlin, Germany * Correspondence: [email protected] Tel.: +49-331-5699-900 References of Figure 1 1Abt et al. 2010, 2Parizzi et al. 2012, 3Hahnke et al. 2016 and Tomazetto et al. 2018, 4Ueki et al. 2006 and Gronow et al. 2011, 5Grabowski et al. 2005, 6Chen and Dong 2005, 7Avgustin et al. 1997 and Purushe et al. 2010, 8Yamada et al. 2006 and Matsuura et al. 2015, 9Yamada et al. 2007 and Matsuura et al. 2015, 10Sun et al. 2016, 11Suen et al. 2011, 12Hahnke et al. 2014 and Tomazetto et al. 2016, 13Mechichi et al. 1999, 14Koeck et al. 2015a and 2015b, 15Tomazetto et al. 2017, 16Fonknechten et al. 2010, 17Chen et al. 2010, 18Nishiyama et al. 2009, 19Sieber et al. 2010, 20Plerce et al. 2008, 21Westerholm et al. 2011 and Müller et al. 2015, 22Ueki et al. 2014, 23Jackson et al. 1999 and McInerney et al. 2007, 24Ma et al. 2017, 25Harmsen et al. 1998 and Plugge et al. 2012, 26Menes and Muxi 2002, Mavromatis et al. 2013 and Hania et al. -

Lascolabacillus Massiliensis'': a New Species Isolated
NEW MICROBES IN HUMANS “Lascolabacillus massiliensis”: a new consent, and the agreement of the National Ethics Committee of Senegal and the local ethics committee of the IFR48 species isolated from the human gut (Marseille, France) were obtained under numbers 11-017 and 09-022, respectively. The initial growth was obtained after 10 days of culture in a 5% sheep blood-enriched sheep rumen M. Beye, S. Bakour, S. I. Traore, D. Raoult and P.-E. Fournier medium in aerobic atmosphere at 37°C. The bacterium was Unité de Recherche en Maladies Infectieuses et Tropicales Emergentes, sub-cultured on 5% sheep blood-enriched Columbia agar Institut Hospitalo-Universitaire Méditerranée-Infection, Aix-Marseille (bioMérieux, Marcy l’Etoile, France) and grew in 24 hours at Université, Faculté de Médecine, Marseille cedex 5, France 37°C in both aerobic and anaerobic conditions. Agar-grown colonies were pale grey and 1.5 mm in diameter. Bacterial cells were Gram-negative, rod-shaped and polymorphic, ranginginlengthfrom1.5to10μm. Strain SIT8 was catalase- Abstract and oxidase-negative. The 16S rRNA gene was sequenced using the fD1-rP2 primers as previously described, using a 3130-XL sequencer (Applied Biosciences, Saint Aubin, We report here the main characteristics of “Lascolabacillus France). Strain SIT8 exhibited a 94.14% sequence identity with massiliensis” strain SIT8 (CSUR P1560) that was isolated from the Proteiniphilum acetatigenes strain TB107 (GenBank accession stool of a healthy 28-month-old boy. NR_043154), the phylogenetically closest species with New Microbes and New Infections © 2016 The Authors. Published standing in nomenclature (Fig. 1), which putatively classifies it by Elsevier Ltd on behalf of European Society of Clinical as a member of a new genus within the family Porphyr- Microbiology and Infectious Diseases. -
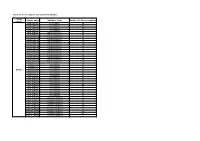
Table S1: List of Samples Included in the Analysis
Table S1: list of samples included in the analysis Study Sample name Inhibitory status Number of days at sampling number DNA.0P2T4 No inhibition 29 DNA.0P2T6 No inhibition 57 DNA.10P2T4 No inhibition 29 DNA.10P2T6 No inhibition 57 DNA.75P2T4 Phenol inhibition 29 DNA.75P2T6 Phenol inhibition 57 DNA.100P2T4 Phenol inhibition 29 DNA.100P2T6 Phenol inhibition 57 DNA.125P1T4 Phenol inhibition 29 DNA.125P1T6 Phenol inhibition 57 DNA.125P2T4 Phenol inhibition 29 DNA.125P2T6 Phenol inhibition 57 DNA.125P3T4 Phenol inhibition 29 DNA.125P3T6 Phenol inhibition 57 DNA.150P2T4 Phenol inhibition 29 DNA.150P2T6 Phenol inhibition 57 DNA.200P2T4 Phenol inhibition 29 DNA.200P2T6 Phenol inhibition 57 DNA.0N2T4 No inhibition 29 DNA.0N2T5 No inhibition 42 DNA.0N2T6 No inhibition 57 Study 1 DNA.5N2T4 No inhibition 29 DNA.5N2T5 No inhibition 42 DNA.5N2T6 No inhibition 57 DNA.10N2T4 No inhibition 29 DNA.10N2T5 No inhibition 42 DNA.10N2T6 No inhibition 57 DNA.15N2T4 No inhibition 29 DNA.15N2T5 No inhibition 42 DNA.15N2T6 No inhibition 57 DNA.25N2T4 No inhibition 29 DNA.25N2T5 No inhibition 42 DNA.25N2T6 No inhibition 57 DNA.75N2T4 Ammonia inhibition 29 DNA.75N2T5 Ammonia inhibition 42 DNA.75N2T6 Ammonia inhibition 57 DNA.100N2T4 Ammonia inhibition 29 DNA.100N2T5 Ammonia inhibition 42 DNA.100N2T6 Ammonia inhibition 57 DNA.250N2T4 Ammonia inhibition 29 DNA.250N2T5 Ammonia inhibition 42 DNA.250N2T6 Ammonia inhibition 57 nono2T3 No inhibition 16 noN2T4 Ammonia inhibition 23 noN2T8 Ammonia inhibition 60 noN2T9 Ammonia inhibition 85 noPhi2T4 Phenol inhibition 23 noPhi2T5 -

EXPERIMENTAL STUDIES on FERMENTATIVE FIRMICUTES from ANOXIC ENVIRONMENTS: ISOLATION, EVOLUTION, and THEIR GEOCHEMICAL IMPACTS By
EXPERIMENTAL STUDIES ON FERMENTATIVE FIRMICUTES FROM ANOXIC ENVIRONMENTS: ISOLATION, EVOLUTION, AND THEIR GEOCHEMICAL IMPACTS By JESSICA KEE EUN CHOI A dissertation submitted to the School of Graduate Studies Rutgers, The State University of New Jersey In partial fulfillment of the requirements For the degree of Doctor of Philosophy Graduate Program in Microbial Biology Written under the direction of Nathan Yee And approved by _______________________________________________________ _______________________________________________________ _______________________________________________________ _______________________________________________________ New Brunswick, New Jersey October 2017 ABSTRACT OF THE DISSERTATION Experimental studies on fermentative Firmicutes from anoxic environments: isolation, evolution and their geochemical impacts by JESSICA KEE EUN CHOI Dissertation director: Nathan Yee Fermentative microorganisms from the bacterial phylum Firmicutes are quite ubiquitous in subsurface environments and play an important biogeochemical role. For instance, fermenters have the ability to take complex molecules and break them into simpler compounds that serve as growth substrates for other organisms. The research presented here focuses on two groups of fermentative Firmicutes, one from the genus Clostridium and the other from the class Negativicutes. Clostridium species are well-known fermenters. Laboratory studies done so far have also displayed the capability to reduce Fe(III), yet the mechanism of this activity has not been investigated -

Syntrophic Butyrate and Propionate Oxidation Processes 491
Environmental Microbiology Reports (2010) 2(4), 489–499 doi:10.1111/j.1758-2229.2010.00147.x Minireview Syntrophic butyrate and propionate oxidation processes: from genomes to reaction mechanismsemi4_147 489..499 Nicolai Müller,1† Petra Worm,2† Bernhard Schink,1* a cytoplasmic fumarate reductase to drive energy- Alfons J. M. Stams2 and Caroline M. Plugge2 dependent succinate oxidation. Furthermore, we 1Faculty for Biology, University of Konstanz, D-78457 propose that homologues of the Thermotoga mar- Konstanz, Germany. itima bifurcating [FeFe]-hydrogenase are involved 2Laboratory of Microbiology, Wageningen University, in NADH oxidation by S. wolfei and S. fumaroxidans Dreijenplein 10, 6703 HB Wageningen, the Netherlands. to form hydrogen. Summary Introduction In anoxic environments such as swamps, rice fields In anoxic environments such as swamps, rice paddy fields and sludge digestors, syntrophic microbial communi- and intestines of higher animals, methanogenic commu- ties are important for decomposition of organic nities are important for decomposition of organic matter to matter to CO2 and CH4. The most difficult step is the CO2 and CH4 (Schink and Stams, 2006; Mcinerney et al., fermentative degradation of short-chain fatty acids 2008; Stams and Plugge, 2009). Moreover, they are the such as propionate and butyrate. Conversion of these key biocatalysts in anaerobic bioreactors that are used metabolites to acetate, CO2, formate and hydrogen is worldwide to treat industrial wastewaters and solid endergonic under standard conditions and occurs wastes. Different types of anaerobes have specified only if methanogens keep the concentrations of these metabolic functions in the degradation pathway and intermediate products low. Butyrate and propionate depend on metabolite transfer which is called syntrophy degradation pathways include oxidation steps of (Schink and Stams, 2006). -

Characterization of Cucumber Fermentation Spoilage Bacteria by Enrichment Culture and 16S Rdna Cloning
Characterization of Cucumber Fermentation Spoilage Bacteria by Enrichment Culture and 16S rDNA Cloning Fred Breidt, Eduardo Medina, Doria Wafa, Ilenys P´erez-D´ıaz, Wendy Franco, Hsin-Yu Huang, Suzanne D. Johanningsmeier, and Jae Ho Kim Abstract: Commercial cucumber fermentations are typically carried out in 40000 L fermentation tanks. A secondary fermentation can occur after sugars are consumed that results in the formation of acetic, propionic, and butyric acids, concomitantly with the loss of lactic acid and an increase in pH. Spoilage fermentations can result in significant economic loss for industrial producers. The microbiota that result in spoilage remain incompletely defined. Previous studies have implicated yeasts, lactic acid bacteria, enterobacteriaceae, and Clostridia as having a role in spoilage fermentations. We report that Propionibacterium and Pectinatus isolates from cucumber fermentation spoilage converted lactic acid to propionic acid, increasing pH. The analysis of 16S rDNA cloning libraries confirmed and expanded the knowledge gained from previous studies using classical microbiological methods. Our data show that Gram-negative anaerobic bacteria supersede Gram-positive Fermincutes species after the pH rises from around 3.2 to pH 5, and propionic and butyric acids are produced. Characterization of the spoilage microbiota is an important first step in efforts to prevent cucumber fermentation spoilage. Keywords: pickled vegetables, Pectinatus, Propionibacteria, secondary cucumber fermentation, spoilage M: Food Microbiology Practical Application: An understanding of the microorganisms that cause commercial cucumber fermentation spoilage & Safety may aid in developing methods to prevent the spoilage from occurring. Introduction cucumbers fermented at 2.3% NaCl (Fleming and others 1989). Commercial cucumber fermentations are typically carried out In this fermentation tank, the initial lactic acid fermentation was in large 40000 L outdoor tanks (reviewed by Breidt and others completed within 2 wk, with 1.2% lactic acid formed (pH 3.6) 2007). -

Microbial Communities Associated with Stable Fly (Diptera: Muscidae) Larvae and Their Developmental Substrates Erin Scully USDA-ARS, [email protected]
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Faculty Publications: Department of Entomology Entomology, Department of 2017 Microbial Communities Associated With Stable Fly (Diptera: Muscidae) Larvae and Their Developmental Substrates Erin Scully USDA-ARS, [email protected] Kristina Friesen USDA-ARS, [email protected] Brian Wienhold USDA-ARS, [email protected] Lisa M. Durso USDA-ARS, [email protected] Follow this and additional works at: http://digitalcommons.unl.edu/entomologyfacpub Part of the Entomology Commons Scully, Erin; Friesen, Kristina; Wienhold, Brian; and Durso, Lisa M., "Microbial Communities Associated With Stable Fly (Diptera: Muscidae) Larvae and Their eD velopmental Substrates" (2017). Faculty Publications: Department of Entomology. 502. http://digitalcommons.unl.edu/entomologyfacpub/502 This Article is brought to you for free and open access by the Entomology, Department of at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Faculty Publications: Department of Entomology by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Annals of the Entomological Society of America, 110(1), 2017, 61–72 doi: 10.1093/aesa/saw087 Special Collection: Filth Fly–Microbe Interactions Research article Microbial Communities Associated With Stable Fly (Diptera: Muscidae) Larvae and Their Developmental Substrates Erin Scully,1 Kristina Friesen,2,3 Brian Wienhold,2 and Lisa M. Durso2 1USDA, ARS, Stored Product -

Page 1 of 41 RSC Advances
RSC Advances This is an Accepted Manuscript, which has been through the Royal Society of Chemistry peer review process and has been accepted for publication. Accepted Manuscripts are published online shortly after acceptance, before technical editing, formatting and proof reading. Using this free service, authors can make their results available to the community, in citable form, before we publish the edited article. This Accepted Manuscript will be replaced by the edited, formatted and paginated article as soon as this is available. You can find more information about Accepted Manuscripts in the Information for Authors. Please note that technical editing may introduce minor changes to the text and/or graphics, which may alter content. The journal’s standard Terms & Conditions and the Ethical guidelines still apply. In no event shall the Royal Society of Chemistry be held responsible for any errors or omissions in this Accepted Manuscript or any consequences arising from the use of any information it contains. www.rsc.org/advances Page 1 of 41 RSC Advances 1 Regulation of acidogenic metabolism towards enhanced short chain fatty acids biosynthesis 2 from waste: Metagenomic Profiling 3 4 Omprakash Sarkar, A. Naresh Kumar, Shikha Dahiya, K.Vamshi Krishna, Dileep Kumar 5 Yeruva, S.Venkata Mohan* 6 Bioengineering and Environmental Sciences (BEES), CSIR-Indian Institute of Chemical 7 Technology (CSIR-IICT), Hyderabad 500 007, India 8 *E-mail: [email protected]; Tel: 0091-40-27161765 9 10 Abstract 11 Short chain carboxylic (volatile fatty) acids (VFA) production in mixed microbiomes is majorly 12 limited by the prevalence of the methanogenic bacteria and availability of substrate from waste 13 to the biocatalyst during the fermentation process. -

Genome Diversity of Spore-Forming Firmicutes MICHAEL Y
Genome Diversity of Spore-Forming Firmicutes MICHAEL Y. GALPERIN National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894 ABSTRACT Formation of heat-resistant endospores is a specific Vibrio subtilis (and also Vibrio bacillus), Ferdinand Cohn property of the members of the phylum Firmicutes (low-G+C assigned it to the genus Bacillus and family Bacillaceae, Gram-positive bacteria). It is found in representatives of four specifically noting the existence of heat-sensitive vegeta- different classes of Firmicutes, Bacilli, Clostridia, Erysipelotrichia, tive cells and heat-resistant endospores (see reference 1). and Negativicutes, which all encode similar sets of core sporulation fi proteins. Each of these classes also includes non-spore-forming Soon after that, Robert Koch identi ed Bacillus anthracis organisms that sometimes belong to the same genus or even as the causative agent of anthrax in cattle and the species as their spore-forming relatives. This chapter reviews the endospores as a means of the propagation of this orga- diversity of the members of phylum Firmicutes, its current taxon- nism among its hosts. In subsequent studies, the ability to omy, and the status of genome-sequencing projects for various form endospores, the specific purple staining by crystal subgroups within the phylum. It also discusses the evolution of the violet-iodine (Gram-positive staining, reflecting the pres- Firmicutes from their apparently spore-forming common ancestor ence of a thick peptidoglycan layer and the absence of and the independent loss of sporulation genes in several different lineages (staphylococci, streptococci, listeria, lactobacilli, an outer membrane), and the relatively low (typically ruminococci) in the course of their adaptation to the saprophytic less than 50%) molar fraction of guanine and cytosine lifestyle in a nutrient-rich environment. -

Microbial Communities Mediating Algal Detritus Turnover Under Anaerobic Conditions
Microbial communities mediating algal detritus turnover under anaerobic conditions Jessica M. Morrison1,*, Chelsea L. Murphy1,*, Kristina Baker1, Richard M. Zamor2, Steve J. Nikolai2, Shawn Wilder3, Mostafa S. Elshahed1 and Noha H. Youssef1 1 Department of Microbiology and Molecular Genetics, Oklahoma State University, Stillwater, OK, USA 2 Grand River Dam Authority, Vinita, OK, USA 3 Department of Integrative Biology, Oklahoma State University, Stillwater, OK, USA * These authors contributed equally to this work. ABSTRACT Background. Algae encompass a wide array of photosynthetic organisms that are ubiquitously distributed in aquatic and terrestrial habitats. Algal species often bloom in aquatic ecosystems, providing a significant autochthonous carbon input to the deeper anoxic layers in stratified water bodies. In addition, various algal species have been touted as promising candidates for anaerobic biogas production from biomass. Surprisingly, in spite of its ecological and economic relevance, the microbial community involved in algal detritus turnover under anaerobic conditions remains largely unexplored. Results. Here, we characterized the microbial communities mediating the degradation of Chlorella vulgaris (Chlorophyta), Chara sp. strain IWP1 (Charophyceae), and kelp Ascophyllum nodosum (phylum Phaeophyceae), using sediments from an anaerobic spring (Zodlteone spring, OK; ZDT), sludge from a secondary digester in a local wastewater treatment plant (Stillwater, OK; WWT), and deeper anoxic layers from a seasonally stratified lake -

WO 2014/121298 A2 7 August 2014 (07.08.2014) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2014/121298 A2 7 August 2014 (07.08.2014) P O P C T (51) International Patent Classification: VULIC, Marin; c/o Seres Health, Inc., 161 First Street, A61K 39/02 (2006.01) Suite 1A, Cambridge, MA 02142 (US). (21) International Application Number: (74) Agents: HUBL, Susan, T. et al; Fenwick & West LLP, PCT/US2014/014738 Silicon Valley Center, 801 California Street, Mountain View, CA 94041 (US). (22) International Filing Date: 4 February 2014 (04.02.2014) (81) Designated States (unless otherwise indicated, for every kind of national protection available): AE, AG, AL, AM, English (25) Filing Language: AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, (26) Publication Language: English BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, (30) Priority Data: HN, HR, HU, ID, IL, IN, IR, IS, JP, KE, KG, KN, KP, KR, 61/760,584 4 February 2013 (04.02.2013) US KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, ME, 61/760,585 4 February 2013 (04.02.2013) US MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, 61/760,574 4 February 2013 (04.02.2013) us OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, 61/760,606 4 February 2013 (04.02.2013) us SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, 61/926,918 13 January 2014 (13.01.2014) us TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, (71) Applicant: SERES HEALTH, INC.