Revised Supplement 1: Reference List for Figure 1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Advance View Proofs
Microbes Environ. Vol. XX, No. X, XXX–XXX, XXXX https://www.jstage.jst.go.jp/browse/jsme2 doi:10.1264/jsme2.ME12193 Isolation and Characterization of a Thermophilic, Obligately Anaerobic and Heterotrophic Marine Chloroflexi Bacterium from a Chloroflexi-dominated Microbial Community Associated with a Japanese Shallow Hydrothermal System, and Proposal for Thermomarinilinea lacunofontalis gen. nov., sp. nov. TAKURO NUNOURA1*, MIHO HIRAI1, MASAYUKI MIYAZAKI1, HIROMI KAZAMA1, HIROKO MAKITA1, HISAKO HIRAYAMA1, YASUO FURUSHIMA2, HIROYUKI YAMAMOTO2, HIROYUKI IMACHI1, and KEN TAKAI1 1Subsurface Geobiology & Advanced Research (SUGAR) Project, Extremobiosphere Research Program, Institute of Biogeosciences, Japan Agency for Marine-Earth Science & Technology (JAMSTEC), 2–15 Natsushima-cho, Yokosuka 237–0061, Japan; and 2Marine Biodiversity Research Program, Institute of Biogeosciences, Japan Agency for Marine-Earth Science & Technology (JAMSTEC), 2–15 Natsushima-cho, Yokosuka 237–0061, Japan (Received October 23, 2012—Accepted January 6, 2013—Published online May 11, 2013) A novel marine thermophilic and heterotrophic Anaerolineae bacterium in the phylum Chloroflexi, strain SW7T, was isolated from an in situ colonization system deployed in the main hydrothermal vent of the Taketomi submarine hot spring field located on the southern part of Yaeyama Archipelago, Japan. The microbial community associated with the hydrothermal vent was predominated by thermophilic heterotrophs such as Thermococcaceae and Anaerolineae, and the next dominant population was thermophilic sulfur oxidizers. Both aerobic and anaerobic hydrogenotrophs including methanogens were detected as minor populations. During the culture-dependent viable count analysis in this study, an Anaerolineae strain SW7T was isolated from an enrichment culture at a high dilution rate. Strain SW7T was an obligately anaerobic heterotroph that grew with fermentation and had non-motileProofs thin rods 3.5–16.5 µm in length and 0.2 µm in width constituting multicellular filaments. -

Co-Occurrence Patterns Among Prokaryotes Across an Age Gradient in Pit Mud of Chinese Strong-Flavor Liquor
Canadian Journal of Microbiology Co-occurrence patterns among prokaryotes across an age gradient in pit mud of Chinese strong-flavor liquor Journal: Canadian Journal of Microbiology Manuscript ID cjm-2020-0012.R1 Manuscript Type: Article Date Submitted by the 04-Mar-2020 Author: Complete List of Authors: Zheng, Yan; Zhengzhou University of Light Industry, School of Food and Biological Engineering Hu, Xiaolong; Zhengzhou University of Light Industry Jia, Zhongjun; Institute of Soil Science Chinese Academy of Sciences Bodelier, Paul;Draft Netherlands Institute of Ecology Guo, Zhiying; Institute of Soil Science Chinese Academy of Sciences Zhang, Yong; Zhengzhou University of Light Industry Li, Fangli; Zhengzhou University of Light Industry He, Peixin; Zhengzhou University of Light Industry prokaryotic community, different aged pit mud, co-occurrence patterns, Keyword: 16S rRNA gene, high-throughput sequencing Is the invited manuscript for consideration in a Special Not applicable (regular submission) Issue? : https://mc06.manuscriptcentral.com/cjm-pubs Page 1 of 35 Canadian Journal of Microbiology 1 (a) The title: 2 Co-occurrence patterns among prokaryotes across an age gradient in pit mud of 3 Chinese strong-flavor liquor 4 (b) Names of authors 5 Yan Zheng1, Xiaolong Hu1, Zhongjun Jia2, Paul L.E. Bodelier 3, Zhiying Guo2, Yong 6 Zhang1, Fangli Li1, Peixin He1# 7 (c) Affiliation and address for each authors 8 1School of Food and Biological Engineering, Zhengzhou University of Light Industry, 9 Zhengzhou, 450002, Henan Province, People’s Republic of China 10 2 State Key Laboratory of Soil and DraftSustainable Agriculture, Institute of Soil Science, 11 Chinese Academy of Sciences, Nanjing, 210008, Jiangsu Province, People’s Republic 12 of China 13 3Netherlands Institute of Ecology (NIOO-KNAW), Department of Microbial Ecology, 14 Droevendaalsesteeg 10, 6708 PB, Wageningen, the Netherlands 15 (d) Email address for each author 16 Yan Zheng, [email protected]; Xiaolong Hu, [email protected]; 17 Zhongjun Jia, [email protected]; Paul L.E. -

Lascolabacillus Massiliensis'': a New Species Isolated
NEW MICROBES IN HUMANS “Lascolabacillus massiliensis”: a new consent, and the agreement of the National Ethics Committee of Senegal and the local ethics committee of the IFR48 species isolated from the human gut (Marseille, France) were obtained under numbers 11-017 and 09-022, respectively. The initial growth was obtained after 10 days of culture in a 5% sheep blood-enriched sheep rumen M. Beye, S. Bakour, S. I. Traore, D. Raoult and P.-E. Fournier medium in aerobic atmosphere at 37°C. The bacterium was Unité de Recherche en Maladies Infectieuses et Tropicales Emergentes, sub-cultured on 5% sheep blood-enriched Columbia agar Institut Hospitalo-Universitaire Méditerranée-Infection, Aix-Marseille (bioMérieux, Marcy l’Etoile, France) and grew in 24 hours at Université, Faculté de Médecine, Marseille cedex 5, France 37°C in both aerobic and anaerobic conditions. Agar-grown colonies were pale grey and 1.5 mm in diameter. Bacterial cells were Gram-negative, rod-shaped and polymorphic, ranginginlengthfrom1.5to10μm. Strain SIT8 was catalase- Abstract and oxidase-negative. The 16S rRNA gene was sequenced using the fD1-rP2 primers as previously described, using a 3130-XL sequencer (Applied Biosciences, Saint Aubin, We report here the main characteristics of “Lascolabacillus France). Strain SIT8 exhibited a 94.14% sequence identity with massiliensis” strain SIT8 (CSUR P1560) that was isolated from the Proteiniphilum acetatigenes strain TB107 (GenBank accession stool of a healthy 28-month-old boy. NR_043154), the phylogenetically closest species with New Microbes and New Infections © 2016 The Authors. Published standing in nomenclature (Fig. 1), which putatively classifies it by Elsevier Ltd on behalf of European Society of Clinical as a member of a new genus within the family Porphyr- Microbiology and Infectious Diseases. -
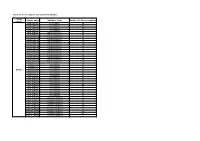
Table S1: List of Samples Included in the Analysis
Table S1: list of samples included in the analysis Study Sample name Inhibitory status Number of days at sampling number DNA.0P2T4 No inhibition 29 DNA.0P2T6 No inhibition 57 DNA.10P2T4 No inhibition 29 DNA.10P2T6 No inhibition 57 DNA.75P2T4 Phenol inhibition 29 DNA.75P2T6 Phenol inhibition 57 DNA.100P2T4 Phenol inhibition 29 DNA.100P2T6 Phenol inhibition 57 DNA.125P1T4 Phenol inhibition 29 DNA.125P1T6 Phenol inhibition 57 DNA.125P2T4 Phenol inhibition 29 DNA.125P2T6 Phenol inhibition 57 DNA.125P3T4 Phenol inhibition 29 DNA.125P3T6 Phenol inhibition 57 DNA.150P2T4 Phenol inhibition 29 DNA.150P2T6 Phenol inhibition 57 DNA.200P2T4 Phenol inhibition 29 DNA.200P2T6 Phenol inhibition 57 DNA.0N2T4 No inhibition 29 DNA.0N2T5 No inhibition 42 DNA.0N2T6 No inhibition 57 Study 1 DNA.5N2T4 No inhibition 29 DNA.5N2T5 No inhibition 42 DNA.5N2T6 No inhibition 57 DNA.10N2T4 No inhibition 29 DNA.10N2T5 No inhibition 42 DNA.10N2T6 No inhibition 57 DNA.15N2T4 No inhibition 29 DNA.15N2T5 No inhibition 42 DNA.15N2T6 No inhibition 57 DNA.25N2T4 No inhibition 29 DNA.25N2T5 No inhibition 42 DNA.25N2T6 No inhibition 57 DNA.75N2T4 Ammonia inhibition 29 DNA.75N2T5 Ammonia inhibition 42 DNA.75N2T6 Ammonia inhibition 57 DNA.100N2T4 Ammonia inhibition 29 DNA.100N2T5 Ammonia inhibition 42 DNA.100N2T6 Ammonia inhibition 57 DNA.250N2T4 Ammonia inhibition 29 DNA.250N2T5 Ammonia inhibition 42 DNA.250N2T6 Ammonia inhibition 57 nono2T3 No inhibition 16 noN2T4 Ammonia inhibition 23 noN2T8 Ammonia inhibition 60 noN2T9 Ammonia inhibition 85 noPhi2T4 Phenol inhibition 23 noPhi2T5 -

An Integrated Study on Microbial Community in Anaerobic Digestion Systems
An Integrated Study on Microbial Community in Anaerobic Digestion Systems DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Yueh-Fen Li Graduate Program in Environmental Science The Ohio State University 2013 Dissertation Committee: Dr. Zhongtang Yu, Advisor Dr. Brian Ahmer Dr. Richard Dick Dr. Olli Tuovinen Copyrighted by Yueh-Fen Li 2013 Abstract Anaerobic digestion (AD) is an attractive microbiological technology for both waste treatment and energy production. Microorganisms are the driving force for the whole transformation process in anaerobic digesters. However, the microbial community underpinning the AD process remains poorly understood, especially with respect to community composition and dynamics in response to variations in feedstocks and operations. The overall objective was to better understand the microbiology driving anaerobic digestion processes by systematically investigating the diversity, composition and succession of microbial communities, both bacterial and archaeal, in anaerobic digesters of different designs, fed different feedstocks, and operated under different conditions. The first two studies focused on propionate-degrading bacteria with an emphasis on syntrophic propionate-oxidizing bacteria. Propionate is one of the most important intermediates and has great influence on AD stability in AD systems because it is inhibitory to methanogens and it can only be metabolized through syntrophic propionate- oxidizing acetogenesis under methanogenic conditions. In the first study (chapter 3), primers specific to the propionate-CoA transferase gene (pct) were designed and used to construct clone libraries, which were sequenced and analyzed to investigate the diversity and distribution of propionate-utilizing bacteria present in the granular and the liquid portions of samples collected from four digesters of different designs, fed different ii feedstocks, and operated at different temperatures. -

Genomics, Exometabolomics, and Metabolic Probing Reveal Conserved Proteolytic Metabolism of Thermoflexus Hugenholtzii and Three Candidate Species from China and Japan
fmicb-12-632731 April 27, 2021 Time: 13:56 # 1 ORIGINAL RESEARCH published: 03 May 2021 doi: 10.3389/fmicb.2021.632731 Genomics, Exometabolomics, and Metabolic Probing Reveal Conserved Proteolytic Metabolism of Thermoflexus hugenholtzii and Three Candidate Species From China and Japan Scott C. Thomas1*, Devon Payne1†, Kevin O. Tamadonfar1†, Cale O. Seymour1, Jian-Yu Jiao2,3, Senthil K. Murugapiran1,4†, Dengxun Lai1, Rebecca Lau5,6, Edited by: Benjamin P. Bowen5,6, Leslie P. Silva5,6, Katherine B. Louie5,6, Marcel Huntemann5,6, Jesse G. Dillon, Alicia Clum5,6, Alex Spunde5,6, Manoj Pillay5,6, Krishnaveni Palaniappan5,6, California State University, Long Neha Varghese5,6, Natalia Mikhailova5,6, I-Min Chen5,6, Dimitrios Stamatis5,6, Beach, United States T. B. K. Reddy5,6, Ronan O’Malley5,6, Chris Daum5,6, Nicole Shapiro5,6, Natalia Ivanova5,6, Reviewed by: Nikos C. Kyrpides5,6, Tanja Woyke5,6, Emiley Eloe-Fadrosh5,6, Trinity L. Hamilton4, Andrew Decker Steen, Paul Dijkstra7, Jeremy A. Dodsworth8, Trent R. Northen5,6, Wen-Jun Li2,3 and The University of Tennessee, Brian P. Hedlund1,9* Knoxville, United States Vera Thiel, 1 School of Life Sciences, University of Nevada, Las Vegas, Las Vegas, NV, United States, 2 School of Life Sciences, Sun German Collection of Microorganisms Yat-sen University, Guangzhou, China, 3 State Key Laboratory of Biocontrol, Guangdong Provincial Key Laboratory of Plant and Cell Cultures GmbH (DSMZ), Resources and Southern Marine Science and Engineering Guangdong Laboratory, Zhuhai, China, 4 Department of Plant Germany and Microbial Biology, The BioTechnology Institute, University of Minnesota, St. Paul, MN, United States, 5 The Department of Energy Joint Genome Institute, Berkeley, CA, United States, 6 Environmental Genomics and Systems Biology Division, *Correspondence: Lawrence Berkeley National Laboratory, Berkeley, CA, United States, 7 Department of Biological Sciences, Center Scott C. -

Syntrophism Among Prokaryotes Bernhard Schink1
Syntrophism Among Prokaryotes Bernhard Schink1 . Alfons J. M. Stams2 1Department of Biology, University of Konstanz, Constance, Germany 2Laboratory of Microbiology, Wageningen University, Wageningen, The Netherlands Introduction: Concepts of Cooperation in Microbial Introduction: Concepts of Cooperation in Communities, Terminology . 471 Microbial Communities, Terminology Electron Flow in Methanogenic and Sulfate-Dependent The study of pure cultures in the laboratory has provided an Degradation . 472 amazingly diverse diorama of metabolic capacities among microorganisms and has established the basis for our under Energetic Aspects . 473 standing of key transformation processes in nature. Pure culture studies are also prerequisites for research in microbial biochem Degradation of Amino Acids . 474 istry and molecular biology. However, desire to understand how Influence of Methanogens . 475 microorganisms act in natural systems requires the realization Obligately Syntrophic Amino Acid Deamination . 475 that microorganisms do not usually occur as pure cultures out Syntrophic Arginine, Threonine, and Lysine there but that every single cell has to cooperate or compete with Fermentation . 475 other micro or macroorganisms. The pure culture is, with some Facultatively Syntrophic Growth with Amino Acids . 476 exceptions such as certain microbes in direct cooperation with Stickland Reaction Versus Methanogenesis . 477 higher organisms, a laboratory artifact. Information gained from the study of pure cultures can be transferred only with Syntrophic Degradation of Fermentation great caution to an understanding of the behavior of microbes in Intermediates . 477 natural communities. Rather, a detailed analysis of the abiotic Syntrophic Ethanol Oxidation . 477 and biotic life conditions at the microscale is needed for a correct Syntrophic Butyrate Oxidation . 478 assessment of the metabolic activities and requirements of Syntrophic Propionate Oxidation . -

The Ecology of the Chloroflexi in Full-Scale Activated Sludge 2 Wastewater Treatment Plants
bioRxiv preprint doi: https://doi.org/10.1101/335752; this version posted May 31, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 The ecology of the Chloroflexi in full-scale activated sludge 2 wastewater treatment plants 3 Marta Nierychlo1, Aleksandra Miłobędzka2,3, Francesca Petriglieri1, Bianca 4 McIlroy1, Per Halkjær Nielsen1, and Simon Jon McIlroy1§* 5 1Center for Microbial Communities, Department of Chemistry and Bioscience, 6 Aalborg University, Aalborg, Denmark 7 2Microbial Ecology and Environmental Biotechnology Department, Institute of 8 Botany, Faculty of Biology, University of Warsaw; Biological and Chemical 9 Research Centre, Żwirki i Wigury 101, Warsaw 02-089, Poland 10 3Department of Biology, Faculty of Building Services, Hydro and Environmental 11 Engineering, Warsaw University of Technology, 00-653 Warsaw, Poland 12 * Corresponding author: Simon Jon McIlroy, Center for Microbial Communities, 13 Department of Chemistry and Bioscience, Aalborg University, Fredrik Bajers Vej 7H, 14 DK-9220 Aalborg, Denmark; Tel.: +45 9940 3573; Fax: +45 9814 1808; Email: 15 [email protected] 16 § Present address: Australian Centre for Ecogenomics, University of Queensland, 17 Australia 1 bioRxiv preprint doi: https://doi.org/10.1101/335752; this version posted May 31, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. -

Characterization of Cucumber Fermentation Spoilage Bacteria by Enrichment Culture and 16S Rdna Cloning
Characterization of Cucumber Fermentation Spoilage Bacteria by Enrichment Culture and 16S rDNA Cloning Fred Breidt, Eduardo Medina, Doria Wafa, Ilenys P´erez-D´ıaz, Wendy Franco, Hsin-Yu Huang, Suzanne D. Johanningsmeier, and Jae Ho Kim Abstract: Commercial cucumber fermentations are typically carried out in 40000 L fermentation tanks. A secondary fermentation can occur after sugars are consumed that results in the formation of acetic, propionic, and butyric acids, concomitantly with the loss of lactic acid and an increase in pH. Spoilage fermentations can result in significant economic loss for industrial producers. The microbiota that result in spoilage remain incompletely defined. Previous studies have implicated yeasts, lactic acid bacteria, enterobacteriaceae, and Clostridia as having a role in spoilage fermentations. We report that Propionibacterium and Pectinatus isolates from cucumber fermentation spoilage converted lactic acid to propionic acid, increasing pH. The analysis of 16S rDNA cloning libraries confirmed and expanded the knowledge gained from previous studies using classical microbiological methods. Our data show that Gram-negative anaerobic bacteria supersede Gram-positive Fermincutes species after the pH rises from around 3.2 to pH 5, and propionic and butyric acids are produced. Characterization of the spoilage microbiota is an important first step in efforts to prevent cucumber fermentation spoilage. Keywords: pickled vegetables, Pectinatus, Propionibacteria, secondary cucumber fermentation, spoilage M: Food Microbiology Practical Application: An understanding of the microorganisms that cause commercial cucumber fermentation spoilage & Safety may aid in developing methods to prevent the spoilage from occurring. Introduction cucumbers fermented at 2.3% NaCl (Fleming and others 1989). Commercial cucumber fermentations are typically carried out In this fermentation tank, the initial lactic acid fermentation was in large 40000 L outdoor tanks (reviewed by Breidt and others completed within 2 wk, with 1.2% lactic acid formed (pH 3.6) 2007). -

Microbial Communities Associated with Stable Fly (Diptera: Muscidae) Larvae and Their Developmental Substrates Erin Scully USDA-ARS, [email protected]
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Faculty Publications: Department of Entomology Entomology, Department of 2017 Microbial Communities Associated With Stable Fly (Diptera: Muscidae) Larvae and Their Developmental Substrates Erin Scully USDA-ARS, [email protected] Kristina Friesen USDA-ARS, [email protected] Brian Wienhold USDA-ARS, [email protected] Lisa M. Durso USDA-ARS, [email protected] Follow this and additional works at: http://digitalcommons.unl.edu/entomologyfacpub Part of the Entomology Commons Scully, Erin; Friesen, Kristina; Wienhold, Brian; and Durso, Lisa M., "Microbial Communities Associated With Stable Fly (Diptera: Muscidae) Larvae and Their eD velopmental Substrates" (2017). Faculty Publications: Department of Entomology. 502. http://digitalcommons.unl.edu/entomologyfacpub/502 This Article is brought to you for free and open access by the Entomology, Department of at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Faculty Publications: Department of Entomology by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Annals of the Entomological Society of America, 110(1), 2017, 61–72 doi: 10.1093/aesa/saw087 Special Collection: Filth Fly–Microbe Interactions Research article Microbial Communities Associated With Stable Fly (Diptera: Muscidae) Larvae and Their Developmental Substrates Erin Scully,1 Kristina Friesen,2,3 Brian Wienhold,2 and Lisa M. Durso2 1USDA, ARS, Stored Product -

Page 1 of 41 RSC Advances
RSC Advances This is an Accepted Manuscript, which has been through the Royal Society of Chemistry peer review process and has been accepted for publication. Accepted Manuscripts are published online shortly after acceptance, before technical editing, formatting and proof reading. Using this free service, authors can make their results available to the community, in citable form, before we publish the edited article. This Accepted Manuscript will be replaced by the edited, formatted and paginated article as soon as this is available. You can find more information about Accepted Manuscripts in the Information for Authors. Please note that technical editing may introduce minor changes to the text and/or graphics, which may alter content. The journal’s standard Terms & Conditions and the Ethical guidelines still apply. In no event shall the Royal Society of Chemistry be held responsible for any errors or omissions in this Accepted Manuscript or any consequences arising from the use of any information it contains. www.rsc.org/advances Page 1 of 41 RSC Advances 1 Regulation of acidogenic metabolism towards enhanced short chain fatty acids biosynthesis 2 from waste: Metagenomic Profiling 3 4 Omprakash Sarkar, A. Naresh Kumar, Shikha Dahiya, K.Vamshi Krishna, Dileep Kumar 5 Yeruva, S.Venkata Mohan* 6 Bioengineering and Environmental Sciences (BEES), CSIR-Indian Institute of Chemical 7 Technology (CSIR-IICT), Hyderabad 500 007, India 8 *E-mail: [email protected]; Tel: 0091-40-27161765 9 10 Abstract 11 Short chain carboxylic (volatile fatty) acids (VFA) production in mixed microbiomes is majorly 12 limited by the prevalence of the methanogenic bacteria and availability of substrate from waste 13 to the biocatalyst during the fermentation process. -

This Is an Open Access-Journal's PDF Published in Chertkov, O., Sikorski
Complete genome sequence of Aminobacterium colombiense type strain (ALA-1T) Authors Chertkov, Olga; Sikorski, Johannes; Brambilla, Evelyne; Lapidus, Alla; Copeland, Alex; Glavina Del Rio, Tijana; Nolan, Matt; Lucas, Susan; Tice, Hope; Cheng, Jan-Fang; Han, Cliff; Detter, John C.; Bruce, David; Tapia, Roxanne; Goodwin, Lynne; Pitluck, Sam; Liolios, Konstantinos; Ivanova, Natalia; Mavromatis, Konstantinos; Ovchinnikova, Galina; Pati, Amrita; Chen, Amy; Palaniappan, Krishna; Land, Miriam; Hauser, Loren; Chang, Yun- Juan; Jeffries, Cynthia D.; Spring, Stefan; Rohde, Manfred; Göker, Markus; Bristow, James; Eisen, Jonathan A.; Markowitz, Victor; Hugenholtz, Philip; Kyrpides, Nikos C.; Klenk, Hans-Peter Citation Complete genome sequence of Aminobacterium colombiense type strain (ALA-1T) 2010, 2 (3):280 Standards in Genomic Sciences DOI 10.4056/sigs.902116 Journal Standards in Genomic Sciences Rights Archived with thanks to Standards in Genomic Sciences Download date 26/09/2021 00:40:08 Link to Item http://hdl.handle.net/2384/297243 This is an Open Access-journal’s PDF published in Chertkov, O., Sikorski, J., Brambilla, E., Lapidus, A., Copeland, A., del Rio, T.G., Nolan, M., Lucas, S., Tice, H., Cheng, J.-F., Han, C., Detter, J.C., Bruce, D., Tapia, R., Goodwin, L., Pitluck, S., Liolios, K., Ivanova, N., Mavromatis, K., Ovchinnikova, G., Pati, A., Chen, A., Palaniappan, K., Land, M., Hauser, L., Chang, Y.-J., Jeffries, C.D., Spring, S., Rohde, M., Göker, M., Bristow, J., Eisen, J.A., Markowitz, V., Hugenholtz, P., Kyrpides, N.C., Klenk, H.-P. Complete genome sequence of Aminobacterium colombiense type strain (ALA-1T) (2010) Standards in Genomic Sciences, 2 (3), pp. 280-289 Standards in Genomic Sciences (2010) 2:280-289 DOI:10.4056/sigs.902116 Complete genome sequence of Aminobacterium T colombiense type strain (ALA-1 ) Olga Chertkov1,2, Johannes Sikorski3, Evelyne Brambilla3, Alla Lapidus1, Alex Copeland1, Tijana Glavina Del Rio1, Matt Nolan1, Susan Lucas1, Hope Tice1, Jan-Fang Cheng1, Cliff Han1,4, John C.