Lascolabacillus Massiliensis'': a New Species Isolated
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

WO 2018/064165 A2 (.Pdf)
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2018/064165 A2 05 April 2018 (05.04.2018) W !P O PCT (51) International Patent Classification: Published: A61K 35/74 (20 15.0 1) C12N 1/21 (2006 .01) — without international search report and to be republished (21) International Application Number: upon receipt of that report (Rule 48.2(g)) PCT/US2017/053717 — with sequence listing part of description (Rule 5.2(a)) (22) International Filing Date: 27 September 2017 (27.09.2017) (25) Filing Language: English (26) Publication Langi English (30) Priority Data: 62/400,372 27 September 2016 (27.09.2016) US 62/508,885 19 May 2017 (19.05.2017) US 62/557,566 12 September 2017 (12.09.2017) US (71) Applicant: BOARD OF REGENTS, THE UNIVERSI¬ TY OF TEXAS SYSTEM [US/US]; 210 West 7th St., Austin, TX 78701 (US). (72) Inventors: WARGO, Jennifer; 1814 Bissonnet St., Hous ton, TX 77005 (US). GOPALAKRISHNAN, Vanch- eswaran; 7900 Cambridge, Apt. 10-lb, Houston, TX 77054 (US). (74) Agent: BYRD, Marshall, P.; Parker Highlander PLLC, 1120 S. Capital Of Texas Highway, Bldg. One, Suite 200, Austin, TX 78746 (US). (81) Designated States (unless otherwise indicated, for every kind of national protection available): AE, AG, AL, AM, AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. -

Revised Supplement 1: Reference List for Figure 1
Revised Supplement 1: Reference list for Figure 1. Manuscript title: Process disturbances in agricultural biogas production – causes, mechanisms and effects on the biogas microbiome: A review Susanne Theuerl 1,*, Johanna Klang 1, Annette Prochnow 1,2 1 Leibniz Institute for Agricultural Engineering and Bioeconomy, Max-Exth-Allee 100, 14469 Potsdam, Germany, [email protected] (ST), [email protected] (JK), [email protected] (AP) 2 Humboldt-Universität zu Berlin, Albrecht-Daniel-Thaer-Institute for Agricultural and Horticultural Sciences, Hinter der Reinhardtstr. 6-8, 10115 Berlin, Germany * Correspondence: [email protected] Tel.: +49-331-5699-900 References of Figure 1 1Abt et al. 2010, 2Parizzi et al. 2012, 3Hahnke et al. 2016 and Tomazetto et al. 2018, 4Ueki et al. 2006 and Gronow et al. 2011, 5Grabowski et al. 2005, 6Chen and Dong 2005, 7Avgustin et al. 1997 and Purushe et al. 2010, 8Yamada et al. 2006 and Matsuura et al. 2015, 9Yamada et al. 2007 and Matsuura et al. 2015, 10Sun et al. 2016, 11Suen et al. 2011, 12Hahnke et al. 2014 and Tomazetto et al. 2016, 13Mechichi et al. 1999, 14Koeck et al. 2015a and 2015b, 15Tomazetto et al. 2017, 16Fonknechten et al. 2010, 17Chen et al. 2010, 18Nishiyama et al. 2009, 19Sieber et al. 2010, 20Plerce et al. 2008, 21Westerholm et al. 2011 and Müller et al. 2015, 22Ueki et al. 2014, 23Jackson et al. 1999 and McInerney et al. 2007, 24Ma et al. 2017, 25Harmsen et al. 1998 and Plugge et al. 2012, 26Menes and Muxi 2002, Mavromatis et al. 2013 and Hania et al. -
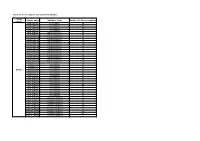
Table S1: List of Samples Included in the Analysis
Table S1: list of samples included in the analysis Study Sample name Inhibitory status Number of days at sampling number DNA.0P2T4 No inhibition 29 DNA.0P2T6 No inhibition 57 DNA.10P2T4 No inhibition 29 DNA.10P2T6 No inhibition 57 DNA.75P2T4 Phenol inhibition 29 DNA.75P2T6 Phenol inhibition 57 DNA.100P2T4 Phenol inhibition 29 DNA.100P2T6 Phenol inhibition 57 DNA.125P1T4 Phenol inhibition 29 DNA.125P1T6 Phenol inhibition 57 DNA.125P2T4 Phenol inhibition 29 DNA.125P2T6 Phenol inhibition 57 DNA.125P3T4 Phenol inhibition 29 DNA.125P3T6 Phenol inhibition 57 DNA.150P2T4 Phenol inhibition 29 DNA.150P2T6 Phenol inhibition 57 DNA.200P2T4 Phenol inhibition 29 DNA.200P2T6 Phenol inhibition 57 DNA.0N2T4 No inhibition 29 DNA.0N2T5 No inhibition 42 DNA.0N2T6 No inhibition 57 Study 1 DNA.5N2T4 No inhibition 29 DNA.5N2T5 No inhibition 42 DNA.5N2T6 No inhibition 57 DNA.10N2T4 No inhibition 29 DNA.10N2T5 No inhibition 42 DNA.10N2T6 No inhibition 57 DNA.15N2T4 No inhibition 29 DNA.15N2T5 No inhibition 42 DNA.15N2T6 No inhibition 57 DNA.25N2T4 No inhibition 29 DNA.25N2T5 No inhibition 42 DNA.25N2T6 No inhibition 57 DNA.75N2T4 Ammonia inhibition 29 DNA.75N2T5 Ammonia inhibition 42 DNA.75N2T6 Ammonia inhibition 57 DNA.100N2T4 Ammonia inhibition 29 DNA.100N2T5 Ammonia inhibition 42 DNA.100N2T6 Ammonia inhibition 57 DNA.250N2T4 Ammonia inhibition 29 DNA.250N2T5 Ammonia inhibition 42 DNA.250N2T6 Ammonia inhibition 57 nono2T3 No inhibition 16 noN2T4 Ammonia inhibition 23 noN2T8 Ammonia inhibition 60 noN2T9 Ammonia inhibition 85 noPhi2T4 Phenol inhibition 23 noPhi2T5 -

Diversity and Composition of the Skin, Blood and Gut Microbiome in Rosacea—A Systematic Review of the Literature
microorganisms Review Diversity and Composition of the Skin, Blood and Gut Microbiome in Rosacea—A Systematic Review of the Literature Klaudia Tutka, Magdalena Zychowska˙ and Adam Reich * Department of Dermatology, Institute of Medical Sciences, Medical College of Rzeszow University, 35-055 Rzeszow, Poland; [email protected] (K.T.); [email protected] (M.Z.)˙ * Correspondence: [email protected]; Tel.: +48-605076722 Received: 30 August 2020; Accepted: 6 November 2020; Published: 8 November 2020 Abstract: Rosacea is a chronic inflammatory skin disorder of a not fully understood pathophysiology. Microbial factors, although not precisely characterized, are speculated to contribute to the development of the condition. The aim of the current review was to summarize the rosacea-associated alterations in the skin, blood, and gut microbiome, investigated using culture-independent, metagenomic techniques. A systematic review of the PubMed, Web of Science, and Scopus databases was performed, according to PRISMA (preferred reporting items for systematic review and meta-analyses) guidelines. Nine out of 185 papers were eligible for analysis. Skin microbiome was investigated in six studies, and in a total number of 115 rosacea patients. Blood microbiome was the subject of one piece of research, conducted in 10 patients with rosacea, and gut microbiome was studied in two papers, and in a total of 23 rosacea subjects. Although all of the studies showed significant alterations in the composition of the skin, blood, or gut microbiome in rosacea, the results were highly inconsistent, or even, in some cases, contradictory. Major limitations included the low number of participants, and different study populations (mainly Asians). Further studies are needed in order to reliably analyze the composition of microbiota in rosacea, and the potential application of microbiome modifications for the treatment of this dermatosis. -

Characterization of Cucumber Fermentation Spoilage Bacteria by Enrichment Culture and 16S Rdna Cloning
Characterization of Cucumber Fermentation Spoilage Bacteria by Enrichment Culture and 16S rDNA Cloning Fred Breidt, Eduardo Medina, Doria Wafa, Ilenys P´erez-D´ıaz, Wendy Franco, Hsin-Yu Huang, Suzanne D. Johanningsmeier, and Jae Ho Kim Abstract: Commercial cucumber fermentations are typically carried out in 40000 L fermentation tanks. A secondary fermentation can occur after sugars are consumed that results in the formation of acetic, propionic, and butyric acids, concomitantly with the loss of lactic acid and an increase in pH. Spoilage fermentations can result in significant economic loss for industrial producers. The microbiota that result in spoilage remain incompletely defined. Previous studies have implicated yeasts, lactic acid bacteria, enterobacteriaceae, and Clostridia as having a role in spoilage fermentations. We report that Propionibacterium and Pectinatus isolates from cucumber fermentation spoilage converted lactic acid to propionic acid, increasing pH. The analysis of 16S rDNA cloning libraries confirmed and expanded the knowledge gained from previous studies using classical microbiological methods. Our data show that Gram-negative anaerobic bacteria supersede Gram-positive Fermincutes species after the pH rises from around 3.2 to pH 5, and propionic and butyric acids are produced. Characterization of the spoilage microbiota is an important first step in efforts to prevent cucumber fermentation spoilage. Keywords: pickled vegetables, Pectinatus, Propionibacteria, secondary cucumber fermentation, spoilage M: Food Microbiology Practical Application: An understanding of the microorganisms that cause commercial cucumber fermentation spoilage & Safety may aid in developing methods to prevent the spoilage from occurring. Introduction cucumbers fermented at 2.3% NaCl (Fleming and others 1989). Commercial cucumber fermentations are typically carried out In this fermentation tank, the initial lactic acid fermentation was in large 40000 L outdoor tanks (reviewed by Breidt and others completed within 2 wk, with 1.2% lactic acid formed (pH 3.6) 2007). -

Microbial Communities Associated with Stable Fly (Diptera: Muscidae) Larvae and Their Developmental Substrates Erin Scully USDA-ARS, [email protected]
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Faculty Publications: Department of Entomology Entomology, Department of 2017 Microbial Communities Associated With Stable Fly (Diptera: Muscidae) Larvae and Their Developmental Substrates Erin Scully USDA-ARS, [email protected] Kristina Friesen USDA-ARS, [email protected] Brian Wienhold USDA-ARS, [email protected] Lisa M. Durso USDA-ARS, [email protected] Follow this and additional works at: http://digitalcommons.unl.edu/entomologyfacpub Part of the Entomology Commons Scully, Erin; Friesen, Kristina; Wienhold, Brian; and Durso, Lisa M., "Microbial Communities Associated With Stable Fly (Diptera: Muscidae) Larvae and Their eD velopmental Substrates" (2017). Faculty Publications: Department of Entomology. 502. http://digitalcommons.unl.edu/entomologyfacpub/502 This Article is brought to you for free and open access by the Entomology, Department of at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Faculty Publications: Department of Entomology by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Annals of the Entomological Society of America, 110(1), 2017, 61–72 doi: 10.1093/aesa/saw087 Special Collection: Filth Fly–Microbe Interactions Research article Microbial Communities Associated With Stable Fly (Diptera: Muscidae) Larvae and Their Developmental Substrates Erin Scully,1 Kristina Friesen,2,3 Brian Wienhold,2 and Lisa M. Durso2 1USDA, ARS, Stored Product -

Uncovering the Potential of Termite Gut Microbiome for Lignocellulose Bioconversion in Anaerobic Batch Bioreactors
ORIGINAL RESEARCH published: 22 December 2017 doi: 10.3389/fmicb.2017.02623 Uncovering the Potential of Termite Gut Microbiome for Lignocellulose Bioconversion in Anaerobic Batch Bioreactors Lucas Auer 1, Adèle Lazuka 1, David Sillam-Dussès 2, 3, Edouard Miambi 4, Michael O’Donohue 1 and Guillermina Hernandez-Raquet 1* 1 Laboratoire d’Ingénierie des Systèmes Biologiques et des Procédés, Université de Toulouse, Centre National de la Recherche Scientifique, Institut National de la Recherche Agronomique, INSA, Toulouse, France, 2 Laboratoire d’Éthologie Expérimentale et Comparée, Université Paris 13 - Sorbonne Paris Cité, Villetaneuse, France, 3 Institut d’Ecologie et des Sciences de l’Environnement de Paris, Institut de Recherche Pour le Développement – Sorbonne Universités, Bondy, France, 4 Institut d’Ecologie et des Sciences de l’Environnement de Paris, Université Paris-Est Créteil, Créteil, France Termites are xylophages, being able to digest a wide variety of lignocellulosic biomass including wood with high lignin content. This ability to feed on recalcitrant plant material Edited by: is the result of complex symbiotic relationships, which involve termite-specific gut Phil B. Pope, microbiomes. Therefore, these represent a potential source of microorganisms for the Norwegian University of Life Sciences, Norway bioconversion of lignocellulose in bioprocesses targeting the production of carboxylates. Reviewed by: In this study, gut microbiomes of four termite species were studied for their capacity to Karen Elizabeth Rossmassler, degrade wheat straw and produce carboxylates in controlled bioreactors. All of the gut Colorado State University, United States microbiomes successfully degraded lignocellulose and up to 45% w/w of wheat straw Zakee L. Sabree, degradation was observed, with the Nasutitermes ephratae gut-microbiome displaying The Ohio State University, the highest levels of wheat straw degradation, carboxylate production and enzymatic United States activity. -

Page 1 of 41 RSC Advances
RSC Advances This is an Accepted Manuscript, which has been through the Royal Society of Chemistry peer review process and has been accepted for publication. Accepted Manuscripts are published online shortly after acceptance, before technical editing, formatting and proof reading. Using this free service, authors can make their results available to the community, in citable form, before we publish the edited article. This Accepted Manuscript will be replaced by the edited, formatted and paginated article as soon as this is available. You can find more information about Accepted Manuscripts in the Information for Authors. Please note that technical editing may introduce minor changes to the text and/or graphics, which may alter content. The journal’s standard Terms & Conditions and the Ethical guidelines still apply. In no event shall the Royal Society of Chemistry be held responsible for any errors or omissions in this Accepted Manuscript or any consequences arising from the use of any information it contains. www.rsc.org/advances Page 1 of 41 RSC Advances 1 Regulation of acidogenic metabolism towards enhanced short chain fatty acids biosynthesis 2 from waste: Metagenomic Profiling 3 4 Omprakash Sarkar, A. Naresh Kumar, Shikha Dahiya, K.Vamshi Krishna, Dileep Kumar 5 Yeruva, S.Venkata Mohan* 6 Bioengineering and Environmental Sciences (BEES), CSIR-Indian Institute of Chemical 7 Technology (CSIR-IICT), Hyderabad 500 007, India 8 *E-mail: [email protected]; Tel: 0091-40-27161765 9 10 Abstract 11 Short chain carboxylic (volatile fatty) acids (VFA) production in mixed microbiomes is majorly 12 limited by the prevalence of the methanogenic bacteria and availability of substrate from waste 13 to the biocatalyst during the fermentation process. -

Proteiniborus Ethanoligenes Gen. Nov., Sp. Nov., an Anaerobic Protein-Utilizing Bacterium
%paper no. ije65108 charlesworth ref: ije65108& New Taxa - Other Gram-positive Bacteria International Journal of Systematic and Evolutionary Microbiology (2008), 58, 000–000 DOI 10.1099/ijs.0.65108-0 Proteiniborus ethanoligenes gen. nov., sp. nov., an anaerobic protein-utilizing bacterium Lili Niu,1,2 Lei Song1,2 and Xiuzhu Dong1 Correspondence 1State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Xiuzhu Dong Sciences, Beijing 100101, PR China [email protected] 2Graduate School, Chinese Academy of Sciences, Beijing 100049, PR China A novel anaerobic, mesophilic, protein-utilizing bacterial strain, GWT, was isolated from the mesophilic hydrogen-producing granular sludge used to treat food industry wastewater. The strain was a Gram-positive, non-spore-forming and non-motile rod. Growth of the strain was observed at 20–48 6C and at pH 6.4–10.0. The strain used yeast extract and peptone as carbon and energy sources. Weak growth was also observed with tryptone and Casamino acids as carbon and energy sources. The strain used none of the tested carbohydrates, alcohols or fatty acids. The fermentation products in peptone-yeast broth included ethanol, acetic acid, hydrogen and carbon dioxide. Gelatin was not hydrolysed. Nitrate was reduced. Indole was produced. T NH3 and H2S were not produced. The DNA G+C content of strain GW was 38.0 mol%. The predominant cellular fatty acids were the saturated fatty acids C14 : 0 (15.58 %), C16 : 0 (25.40 %) and C18 : 0 (12.03 %). Phylogenetic analysis based on 16S rRNA gene sequence similarity revealed that strain GWT represented a new branch within cluster XII of the Clostridium subphylum, with ,89.6 % 16S rRNA gene sequence similarities to all described species. -

Draft Genome Sequence of Fermentimonas Caenicola Strain SIT8, Isolated from the Human
Beye et al. Standards in Genomic Sciences (2018) 13:8 https://doi.org/10.1186/s40793-018-0310-6 EXTENDED GENOME REPORT Open Access Draft genome sequence of Fermentimonas caenicola strain SIT8, isolated from the human gut Mamadou Beye1, Sofiane Bakour1, Sory Ibrahima Traore1, Jaishriram Rathored1, Noémie Labas1, Didier Raoult1,2 and Pierre-Edouard Fournier1* Abstract We report the properties of a draft genome sequence of the bacterium Fermentimonas caenicola strain SIT8 (= CSUR P1560). This strain, whose genome is described here, was isolated from the fecal flora of a healthy 28-month-old Senegalese boy. Strain SIT8 is a facultatively anaerobic Gram-negative bacillus. Here we describe the features of this organism, together with the complete genome sequence and annotation. The 2,824,451-bp long (1 chromosome but no plasmid) contains 2354 protein-coding and 46 RNA genes, including four rRNA genes. Keywords: Fermentimonas caenicola,Genome,Humangut,Microbiota Introduction Herein, we present a set of features for F. caenicola Fermentimonas caenicola strain SIT8 (= CSUR P1560) strain SIT8 together with the description of the was isolated from the stool of a healthy 28-month-old complete genomic sequence and annotation. Senegalese boy as part of a study aiming at cultivating all species within the human gastro-intestinal micro- Organism information biota. It is a Gram-negative, facultatively anaerobic, Classification and features indole-negative bacillus. Initially, we had named this Fermentimonas caenicola “Lascolabacillus massiliensis” strain SIT8 was isolated from bacterium as it exhibited the stool of a healthy 28-month-old Senegalese boy unique features among members of the family Porphyro- ’ monadaceae (Table 1). -

Supplementary Material Evidence of Spatial Homogeneity in an Electromethanogenic Cathodic Microbial Community
Evidence of Spatial Homogeneity in an Electromethanogenic Cathodic Microbial Community. Item Type Article Authors Ragab, Alaa I.; Katuri, Krishna; Ali, Muhammad; Saikaly, Pascal Citation Ragab, A., Katuri, K. P., Ali, M., & Saikaly, P. E. (2019). Evidence of Spatial Homogeneity in an Electromethanogenic Cathodic Microbial Community. Frontiers in Microbiology, 10. doi:10.3389/ fmicb.2019.01747 Eprint version Publisher's Version/PDF DOI 10.3389/fmicb.2019.01747 Publisher Frontiers Media SA Journal Frontiers in microbiology Rights Ragab, Katuri, Ali and Saikaly. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms. Download date 26/09/2021 21:00:48 Item License https://creativecommons.org/licenses/by/4.0/ Link to Item http://hdl.handle.net/10754/656542 Supplementary Material Evidence of spatial homogeneity in an electromethanogenic cathodic microbial community Ala’a Ragab1, Krishna P. Katuri1, Muhammad Ali1, Pascal E. Saikaly1* 1 Biological and Environmental Science and Engineering Division, Water Desalination and Reuse Center, King Abdullah University of Science and Technology (KAUST), Saudi Arabia * Correspondence: Pascal E. Saikaly [email protected] Supplementary Material Figure S1. Rarefaction curves for all retrieved reads for the 263 operational taxonomic units (OTUs) for all samples faceted by reactors and combined together. Data points are colored by sampling position (top, center, bottom). -

Genomic Characterization of the Uncultured Bacteroidales Family S24-7 Inhabiting the Guts of Homeothermic Animals Kate L
Ormerod et al. Microbiome (2016) 4:36 DOI 10.1186/s40168-016-0181-2 RESEARCH Open Access Genomic characterization of the uncultured Bacteroidales family S24-7 inhabiting the guts of homeothermic animals Kate L. Ormerod1, David L. A. Wood1, Nancy Lachner1, Shaan L. Gellatly2, Joshua N. Daly1, Jeremy D. Parsons3, Cristiana G. O. Dal’Molin4, Robin W. Palfreyman4, Lars K. Nielsen4, Matthew A. Cooper5, Mark Morrison6, Philip M. Hansbro2 and Philip Hugenholtz1* Abstract Background: Our view of host-associated microbiota remains incomplete due to the presence of as yet uncultured constituents. The Bacteroidales family S24-7 is a prominent example of one of these groups. Marker gene surveys indicate that members of this family are highly localized to the gastrointestinal tracts of homeothermic animals and are increasingly being recognized as a numerically predominant member of the gut microbiota; however, little is known about the nature of their interactions with the host. Results: Here, we provide the first whole genome exploration of this family, for which we propose the name “Candidatus Homeothermaceae,” using 30 population genomes extracted from fecal samples of four different animal hosts: human, mouse, koala, and guinea pig. We infer the core metabolism of “Ca. Homeothermaceae” to be that of fermentative or nanaerobic bacteria, resembling that of related Bacteroidales families. In addition, we describe three trophic guilds within the family, plant glycan (hemicellulose and pectin), host glycan, and α-glucan, each broadly defined by increased abundance of enzymes involved in the degradation of particular carbohydrates. Conclusions: “Ca. Homeothermaceae” representatives constitute a substantial component of the murine gut microbiota, as well as being present within the human gut, and this study provides important first insights into the nature of their residency.