Microbial Communities Associated with Stable Fly (Diptera: Muscidae) Larvae and Their Developmental Substrates Erin Scully USDA-ARS, [email protected]
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Medical and Veterinary Entomology (2009) 23 (Suppl
Medical and Veterinary Entomology (2009) 23 (Suppl. 1), 1–7 Enabling technologies to improve area-wide integrated pest management programmes for the control of screwworms A. S. ROBINSON , M. J. B. VREYSEN , J. HENDRICHS and U. FELDMANN Joint Food and Agriculture Organization of the United Nations/International Atomic Energy Agency (FAO/IAEA) Programme of Nuclear Techniques in Food and Agriculture, Vienna, Austria Abstract . The economic devastation caused in the past by the New World screwworm fly Cochliomyia hominivorax (Coquerel) (Diptera: Calliphoridae) to the livestock indus- try in the U.S.A., Mexico and the rest of Central America was staggering. The eradication of this major livestock pest from North and Central America using the sterile insect tech- nique (SIT) as part of an area-wide integrated pest management (AW-IPM) programme was a phenomenal technical and managerial accomplishment with enormous economic implications. The area is maintained screwworm-free by the weekly release of 40 million sterile flies in the Darien Gap in Panama, which prevents migration from screwworm- infested areas in Columbia. However, the species is still a major pest in many areas of the Caribbean and South America and there is considerable interest in extending the eradica- tion programme to these countries. Understanding New World screwworm fly popula- tions in the Caribbean and South America, which represent a continuous threat to the screwworm-free areas of Central America and the U.S.A., is a prerequisite to any future eradication campaigns. The Old World screwworm fly Chrysomya bezziana Villeneuve (Diptera: Calliphoridae) has a very wide distribution ranging from Southern Africa to Papua New Guinea and, although its economic importance is assumed to be less than that of its New World counterpart, it is a serious pest in extensive livestock production and a constant threat to pest-free areas such as Australia. -

Revised Supplement 1: Reference List for Figure 1
Revised Supplement 1: Reference list for Figure 1. Manuscript title: Process disturbances in agricultural biogas production – causes, mechanisms and effects on the biogas microbiome: A review Susanne Theuerl 1,*, Johanna Klang 1, Annette Prochnow 1,2 1 Leibniz Institute for Agricultural Engineering and Bioeconomy, Max-Exth-Allee 100, 14469 Potsdam, Germany, [email protected] (ST), [email protected] (JK), [email protected] (AP) 2 Humboldt-Universität zu Berlin, Albrecht-Daniel-Thaer-Institute for Agricultural and Horticultural Sciences, Hinter der Reinhardtstr. 6-8, 10115 Berlin, Germany * Correspondence: [email protected] Tel.: +49-331-5699-900 References of Figure 1 1Abt et al. 2010, 2Parizzi et al. 2012, 3Hahnke et al. 2016 and Tomazetto et al. 2018, 4Ueki et al. 2006 and Gronow et al. 2011, 5Grabowski et al. 2005, 6Chen and Dong 2005, 7Avgustin et al. 1997 and Purushe et al. 2010, 8Yamada et al. 2006 and Matsuura et al. 2015, 9Yamada et al. 2007 and Matsuura et al. 2015, 10Sun et al. 2016, 11Suen et al. 2011, 12Hahnke et al. 2014 and Tomazetto et al. 2016, 13Mechichi et al. 1999, 14Koeck et al. 2015a and 2015b, 15Tomazetto et al. 2017, 16Fonknechten et al. 2010, 17Chen et al. 2010, 18Nishiyama et al. 2009, 19Sieber et al. 2010, 20Plerce et al. 2008, 21Westerholm et al. 2011 and Müller et al. 2015, 22Ueki et al. 2014, 23Jackson et al. 1999 and McInerney et al. 2007, 24Ma et al. 2017, 25Harmsen et al. 1998 and Plugge et al. 2012, 26Menes and Muxi 2002, Mavromatis et al. 2013 and Hania et al. -

Lascolabacillus Massiliensis'': a New Species Isolated
NEW MICROBES IN HUMANS “Lascolabacillus massiliensis”: a new consent, and the agreement of the National Ethics Committee of Senegal and the local ethics committee of the IFR48 species isolated from the human gut (Marseille, France) were obtained under numbers 11-017 and 09-022, respectively. The initial growth was obtained after 10 days of culture in a 5% sheep blood-enriched sheep rumen M. Beye, S. Bakour, S. I. Traore, D. Raoult and P.-E. Fournier medium in aerobic atmosphere at 37°C. The bacterium was Unité de Recherche en Maladies Infectieuses et Tropicales Emergentes, sub-cultured on 5% sheep blood-enriched Columbia agar Institut Hospitalo-Universitaire Méditerranée-Infection, Aix-Marseille (bioMérieux, Marcy l’Etoile, France) and grew in 24 hours at Université, Faculté de Médecine, Marseille cedex 5, France 37°C in both aerobic and anaerobic conditions. Agar-grown colonies were pale grey and 1.5 mm in diameter. Bacterial cells were Gram-negative, rod-shaped and polymorphic, ranginginlengthfrom1.5to10μm. Strain SIT8 was catalase- Abstract and oxidase-negative. The 16S rRNA gene was sequenced using the fD1-rP2 primers as previously described, using a 3130-XL sequencer (Applied Biosciences, Saint Aubin, We report here the main characteristics of “Lascolabacillus France). Strain SIT8 exhibited a 94.14% sequence identity with massiliensis” strain SIT8 (CSUR P1560) that was isolated from the Proteiniphilum acetatigenes strain TB107 (GenBank accession stool of a healthy 28-month-old boy. NR_043154), the phylogenetically closest species with New Microbes and New Infections © 2016 The Authors. Published standing in nomenclature (Fig. 1), which putatively classifies it by Elsevier Ltd on behalf of European Society of Clinical as a member of a new genus within the family Porphyr- Microbiology and Infectious Diseases. -

Evolutionary Background Entities at the Cellular and Subcellular Levels in Bodies of Invertebrate Animals
The Journal of Theoretical Fimpology Volume 2, Issue 4: e-20081017-2-4-14 December 28, 2014 www.fimpology.com Evolutionary Background Entities at the Cellular and Subcellular Levels in Bodies of Invertebrate Animals Shu-dong Yin Cory H. E. R. & C. Inc. Burnaby, British Columbia, Canada Email: [email protected] ________________________________________________________________________ Abstract The novel recognition that individual bodies of normal animals are actually inhabited by subcellular viral entities and membrane-enclosed microentities, prokaryotic bacterial and archaeal cells and unicellular eukaryotes such as fungi and protists has been supported by increasing evidences since the emergence of culture-independent approaches. However, how to understand the relationship between animal hosts including human beings and those non-host microentities or microorganisms is challenging our traditional understanding of pathogenic relationship in human medicine and veterinary medicine. In recent novel evolution theories, the relationship between animals and their environments has been deciphered to be the interaction between animals and their environmental evolutionary entities at the same and/or different evolutionary levels;[1-3] and evolutionary entities of the lower evolutionary levels are hypothesized to be the evolutionary background entities of entities at the higher evolutionary levels.[1,2] Therefore, to understand the normal existence of microentities or microorganisms in multicellular animal bodies is becoming the first priority for elucidating the ecological and evolutiological relationships between microorganisms and nonhuman macroorganisms. The evolutionary background entities at the cellular and subcellular levels in bodies of nonhuman vertebrate animals have been summarized recently.[4] In this paper, the author tries to briefly review the evolutionary background entities (EBE) at the cellular and subcellular levels for several selected invertebrate animal species. -
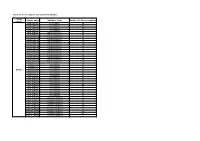
Table S1: List of Samples Included in the Analysis
Table S1: list of samples included in the analysis Study Sample name Inhibitory status Number of days at sampling number DNA.0P2T4 No inhibition 29 DNA.0P2T6 No inhibition 57 DNA.10P2T4 No inhibition 29 DNA.10P2T6 No inhibition 57 DNA.75P2T4 Phenol inhibition 29 DNA.75P2T6 Phenol inhibition 57 DNA.100P2T4 Phenol inhibition 29 DNA.100P2T6 Phenol inhibition 57 DNA.125P1T4 Phenol inhibition 29 DNA.125P1T6 Phenol inhibition 57 DNA.125P2T4 Phenol inhibition 29 DNA.125P2T6 Phenol inhibition 57 DNA.125P3T4 Phenol inhibition 29 DNA.125P3T6 Phenol inhibition 57 DNA.150P2T4 Phenol inhibition 29 DNA.150P2T6 Phenol inhibition 57 DNA.200P2T4 Phenol inhibition 29 DNA.200P2T6 Phenol inhibition 57 DNA.0N2T4 No inhibition 29 DNA.0N2T5 No inhibition 42 DNA.0N2T6 No inhibition 57 Study 1 DNA.5N2T4 No inhibition 29 DNA.5N2T5 No inhibition 42 DNA.5N2T6 No inhibition 57 DNA.10N2T4 No inhibition 29 DNA.10N2T5 No inhibition 42 DNA.10N2T6 No inhibition 57 DNA.15N2T4 No inhibition 29 DNA.15N2T5 No inhibition 42 DNA.15N2T6 No inhibition 57 DNA.25N2T4 No inhibition 29 DNA.25N2T5 No inhibition 42 DNA.25N2T6 No inhibition 57 DNA.75N2T4 Ammonia inhibition 29 DNA.75N2T5 Ammonia inhibition 42 DNA.75N2T6 Ammonia inhibition 57 DNA.100N2T4 Ammonia inhibition 29 DNA.100N2T5 Ammonia inhibition 42 DNA.100N2T6 Ammonia inhibition 57 DNA.250N2T4 Ammonia inhibition 29 DNA.250N2T5 Ammonia inhibition 42 DNA.250N2T6 Ammonia inhibition 57 nono2T3 No inhibition 16 noN2T4 Ammonia inhibition 23 noN2T8 Ammonia inhibition 60 noN2T9 Ammonia inhibition 85 noPhi2T4 Phenol inhibition 23 noPhi2T5 -

Characterization of Cucumber Fermentation Spoilage Bacteria by Enrichment Culture and 16S Rdna Cloning
Characterization of Cucumber Fermentation Spoilage Bacteria by Enrichment Culture and 16S rDNA Cloning Fred Breidt, Eduardo Medina, Doria Wafa, Ilenys P´erez-D´ıaz, Wendy Franco, Hsin-Yu Huang, Suzanne D. Johanningsmeier, and Jae Ho Kim Abstract: Commercial cucumber fermentations are typically carried out in 40000 L fermentation tanks. A secondary fermentation can occur after sugars are consumed that results in the formation of acetic, propionic, and butyric acids, concomitantly with the loss of lactic acid and an increase in pH. Spoilage fermentations can result in significant economic loss for industrial producers. The microbiota that result in spoilage remain incompletely defined. Previous studies have implicated yeasts, lactic acid bacteria, enterobacteriaceae, and Clostridia as having a role in spoilage fermentations. We report that Propionibacterium and Pectinatus isolates from cucumber fermentation spoilage converted lactic acid to propionic acid, increasing pH. The analysis of 16S rDNA cloning libraries confirmed and expanded the knowledge gained from previous studies using classical microbiological methods. Our data show that Gram-negative anaerobic bacteria supersede Gram-positive Fermincutes species after the pH rises from around 3.2 to pH 5, and propionic and butyric acids are produced. Characterization of the spoilage microbiota is an important first step in efforts to prevent cucumber fermentation spoilage. Keywords: pickled vegetables, Pectinatus, Propionibacteria, secondary cucumber fermentation, spoilage M: Food Microbiology Practical Application: An understanding of the microorganisms that cause commercial cucumber fermentation spoilage & Safety may aid in developing methods to prevent the spoilage from occurring. Introduction cucumbers fermented at 2.3% NaCl (Fleming and others 1989). Commercial cucumber fermentations are typically carried out In this fermentation tank, the initial lactic acid fermentation was in large 40000 L outdoor tanks (reviewed by Breidt and others completed within 2 wk, with 1.2% lactic acid formed (pH 3.6) 2007). -

A Case in a Puppy and Overview of Geographical Distribution
ACTA VET. BRNO 2020, 89: 171–177; https://doi.org/10.2754/avb202089020171 Wohlfahrtiosis in Italy: a case in a puppy and overview of geographical distribution Teresa Bonacci1, Giuseppe Curia2, Chiara Scapoli3, Marco Pezzi3 1University of Calabria, Department of Biology, Ecology and Earth Science, Cosenza, Italy 2Azienda Sanitaria Provinciale di Cosenza, Servizio Veterinario, Cosenza, Italy 3University of Ferrara, Department of Life Sciences and Biotechnology, Ferrara, Italy Received November 26, 2019 Accepted April 30, 2020 Abstract The report describes a case of urogenital myiasis in a puppy, Canis lupus familiaris (Carnivora: Canidae) caused by Wohlfahrtia magnifica (Diptera: Sarcophagidae) in Calabria, southern Italy. This species is an obligatory agent of myiasis in human and other warm-blooded vertebrates. The puppy was healthy and was not living near farm animals, usual hosts of this flesh fly. An overview of cases of human and animal myiasis caused by W. magnifica in Italy and of data and specimens documented in entomology museum collections is also reported. Canine, urogenital myiasis, Wohlfahrtia magnifica Myiasis is an important parasitic disease caused by larvae of Diptera infesting vertebrates actively feeding on host tissues (Zumpt 1965). The term “wohlfahrtiosis” refers to myiasis caused by Wohlfahrtia magnifica (Schiner, 1862) (Insecta: Diptera: Sarcophagidae). Among the types of myiasis, wohlfahrtiosis is especially important not only because it may affect humans, but also because it usually induces serious damage due to the high number of deposited larvae and to their rapid growth. When attacking livestock, the parasite may cause heavy economic damages through loss of production and death (Hall and Farkas 2000). In Europe wohlfahrtiosis is an infestation reported in humans and domestic animals in several countries, especially in southern and eastern areas. -

Page 1 of 41 RSC Advances
RSC Advances This is an Accepted Manuscript, which has been through the Royal Society of Chemistry peer review process and has been accepted for publication. Accepted Manuscripts are published online shortly after acceptance, before technical editing, formatting and proof reading. Using this free service, authors can make their results available to the community, in citable form, before we publish the edited article. This Accepted Manuscript will be replaced by the edited, formatted and paginated article as soon as this is available. You can find more information about Accepted Manuscripts in the Information for Authors. Please note that technical editing may introduce minor changes to the text and/or graphics, which may alter content. The journal’s standard Terms & Conditions and the Ethical guidelines still apply. In no event shall the Royal Society of Chemistry be held responsible for any errors or omissions in this Accepted Manuscript or any consequences arising from the use of any information it contains. www.rsc.org/advances Page 1 of 41 RSC Advances 1 Regulation of acidogenic metabolism towards enhanced short chain fatty acids biosynthesis 2 from waste: Metagenomic Profiling 3 4 Omprakash Sarkar, A. Naresh Kumar, Shikha Dahiya, K.Vamshi Krishna, Dileep Kumar 5 Yeruva, S.Venkata Mohan* 6 Bioengineering and Environmental Sciences (BEES), CSIR-Indian Institute of Chemical 7 Technology (CSIR-IICT), Hyderabad 500 007, India 8 *E-mail: [email protected]; Tel: 0091-40-27161765 9 10 Abstract 11 Short chain carboxylic (volatile fatty) acids (VFA) production in mixed microbiomes is majorly 12 limited by the prevalence of the methanogenic bacteria and availability of substrate from waste 13 to the biocatalyst during the fermentation process. -

Human Urinary Myiasis Due to Larvae of Clogmia (Telmatoscopus) Albipunctata Williston (Diptera: Psychodidae) First Report in Egypt
J Vector Borne Dis 51, September 2014, pp. 247–249 Human urinary myiasis due to larvae of Clogmia (Telmatoscopus) albipunctata Williston (Diptera: Psychodidae) first report in Egypt Ayman A. El-Badry1, Hosni Khairy Salem2, Yusuf Abd El-Aziz Edmardash3 1Medical Parasitology Department; 2Urology Department, Kasr Al-Ainy Faculty of Medicine; 3Entomology Department, Faculty of Science, Cairo University, Cairo, Egypt Key words Clogmia albipunctata; Egypt; human myasis Human myiasis is defined as “the infestation of the Case report tissue of living human with dipterous larvae”1. Parasito- The patient presented with repeated passage of nu- logically myiasis could be classified as obligatory, facul- merous living dark-colored larvae in urine, 7–12 larvae tative or accidental. Clinically myiasis may be classified were voided intermittently over two months. She was according to part of the body tissue invaded. Cutaneous complaining of dysuria, fever and itching in the periure- myiasis is the commonest type. Body cavity myiasis; na- thral and genital regions. No history of travelling outside sopharyngeal, ocular, aural and the gastrointestinal tract Egypt in the past or the present time. Complete urine urogenital system are less common. Urethral myiasis is analysis and stool examination using direct and concen- exceptionally rare, even in sites usually protected by trated smear was done. Plain X-ray and pelviabdominal clothes, inaccessible for the flies1–2. A large number of ultrasound were also done. fly species may cause urinary myiasis. Larvae of Fannia Larvae from two different fresh urine samples were scalaris3 is the most frequent cause of urinary myiasis. identified morphologically as larvae of Clogmia Other fly genera Musca, Sarcophaga, Lucilia, Wohlfahr- albipunctata (Diptera: Psychodidae). -

XIII) Arribas, Manuela Guerrero, José Mª Prieto, Mª Pilar Rodríguez E Isabel Morón) Y Del Servicio De J
13. Nuevas_2009 7/1/10 12:15 Página 249 Graellsia, 65(2): 249-280 (2009) NOTICIA DE NUEVOS TÁXONES PARA LA CIENCIA EN EL ÁMBITO ÍBERO-BALEAR Y MACARONÉSICO Nuevos táxones animales descritos en la península Museo Nacional de Ciencias Naturales (Purificación Ibérica y Macaronesia desde 1994 (XIII) Arribas, Manuela Guerrero, José Mª Prieto, Mª Pilar Rodríguez e Isabel Morón) y del Servicio de J. FERNÁNDEZ Museo Nacional de Ciencias Naturales, C.S.I.C. Reprografía (Ana Aguilar), así como de Rafael Araujo José Gutiérrez Abascal, 2. 28006. Madrid. (bibliotecario de la Sociedad Española de E-mail: [email protected] Malacología). Como en otras ocasiones, el Proyecto Fauna Ibérica IX (CGL2007-66786-CO8-01) subvenciona la Otra vez al final del año presentamos una nueva edición de estas notas y el apoyo de las editoras y del relación de novedades taxonómicas en el ámbito ibé- Comité de Redacción de Graellsia las hace posible. rico y macaronésico. Las características de esta nueva entrega son las habituales: se incluyen todos aquéllos táxones de nueva descripción que incluyan represen- tantes en el área considerada; en el caso de las espe- cies sólo se consideran aquéllas cuya localidad tipo se *Kryptrochozoa Dunn, Edgecombe, Giribet, Hejnol, Martindale y Rouse, 2009 encuentra en el área considerada y las que entre su REFERENCIA: Giribet, G., Dunn, C.W., Edgecombe, G.D., Hejnol, A., distribución conocida en el momento de la descrip- Rouse, G.W. y Martindale, M.Q., 2009. Assembling the spiralian ción haya alguna parte de la zona considerada. El tree of life. En: Animal evolution: genomes, fossils, and trees. -

Proteiniborus Ethanoligenes Gen. Nov., Sp. Nov., an Anaerobic Protein-Utilizing Bacterium
%paper no. ije65108 charlesworth ref: ije65108& New Taxa - Other Gram-positive Bacteria International Journal of Systematic and Evolutionary Microbiology (2008), 58, 000–000 DOI 10.1099/ijs.0.65108-0 Proteiniborus ethanoligenes gen. nov., sp. nov., an anaerobic protein-utilizing bacterium Lili Niu,1,2 Lei Song1,2 and Xiuzhu Dong1 Correspondence 1State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Xiuzhu Dong Sciences, Beijing 100101, PR China [email protected] 2Graduate School, Chinese Academy of Sciences, Beijing 100049, PR China A novel anaerobic, mesophilic, protein-utilizing bacterial strain, GWT, was isolated from the mesophilic hydrogen-producing granular sludge used to treat food industry wastewater. The strain was a Gram-positive, non-spore-forming and non-motile rod. Growth of the strain was observed at 20–48 6C and at pH 6.4–10.0. The strain used yeast extract and peptone as carbon and energy sources. Weak growth was also observed with tryptone and Casamino acids as carbon and energy sources. The strain used none of the tested carbohydrates, alcohols or fatty acids. The fermentation products in peptone-yeast broth included ethanol, acetic acid, hydrogen and carbon dioxide. Gelatin was not hydrolysed. Nitrate was reduced. Indole was produced. T NH3 and H2S were not produced. The DNA G+C content of strain GW was 38.0 mol%. The predominant cellular fatty acids were the saturated fatty acids C14 : 0 (15.58 %), C16 : 0 (25.40 %) and C18 : 0 (12.03 %). Phylogenetic analysis based on 16S rRNA gene sequence similarity revealed that strain GWT represented a new branch within cluster XII of the Clostridium subphylum, with ,89.6 % 16S rRNA gene sequence similarities to all described species. -

Cutaneous Myiasis in a Child Scalp Caused by Wohlfahrtia Magnifica (Diptera: Sarcophagidae): a Case Report
MOJ Clinical & Medical Case Reports Case Report Open Access Cutaneous myiasis in a child scalp caused by Wohlfahrtia Magnifica (Diptera: sarcophagidae): a case report Abstract Volume 4 Issue 3 - 2016 Background: Myiasis is caused by the invasion of tissues or organs of man or animals Raghad Al Badri,1 Taiba Al Harbi,1 Assad by dipterous larvae. A four-year-old girl presented with one month history of scalp 1 2 3,4 ulcer that has initially started as a painful itchy swelling in the occipital region. Tonnsi, Amal Almatary, Raafat Hassanein 1Pediatric Department, Maternity and Children Hospital, Saudi Physical examination revealed live maggots in the ulcerous wound. The maggots were Arabia identified as the third instar larvae of Wohlfahrtia magnifica. 2Department of Parasitology, Assiut University, Egypt 3 Case presentation: The patient presented to the emergency department with larva Department of Laboratory Medicine, Umm Al-Qura University, visualized inside the ulcer by the parents, the clinical examination has revealed a well Saudi Arabia 4Department of Animal Hygiene and Zoonoses, Assiut circumscribed circular lesion in the occipital region of the scalp with a diameter of University, Egypt approximately 4cm, it showed some signs of inflammation with necrotic tissue and many larva’s were seen inside the ulcer. The first surgical debridement was done in Correspondence: Raafat Hassanein, Department of the second day of admission to the hospital, during this operation a large number of Laboratory Medicine, Faculty of Applied Medical Sciences, Umm larvae were extracted and larval specimens were identified morphologically as larvae Al-Qura University, Makkah, Saudi Arabia, Tel +966-531664354, of Wohlfahrtia magnifica (Diptera: Sarcophagidae).