Assessing Model Sensitivity in Ancestral Area Reconstruction Using
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Plant List 2016
Established 1990 PLANT LIST 2016 European mail order website www.crug-farm.co.uk CRÛG FARM PLANTS • 2016 Welcome to our 2016 list hope we can tempt you with plenty of our old favourites as well as some exciting new plants that we have searched out on our travels. There has been little chance of us standing still with what has been going on here in 2015. The year started well with the birth of our sixth grandchild. January into February had Sue and I in Colombia for our first winter/early spring expedition. It was exhilarating, we were able to travel much further afield than we had previously, as the mountainous areas become safer to travel. We are looking forward to working ever closer with the Colombian institutes, such as the Medellin Botanic Gardens whom we met up with. Consequently we were absent from the RHS February Show at Vincent Square. We are finding it increasingly expensive participating in the London shows, while re-branding the RHS February Show as a potato event hardly encourages our type of customer base to visit. A long standing speaking engagement and a last minute change of date, meant that we missed going to Fota near Cork last spring, no such problem this coming year. We were pleasantly surprised at the level of interest at the Trgrehan Garden Rare Plant Fair, in Cornwall. Hopefully this will become an annual event for us, as well as the Cornwall Garden Society show in April. Poor Sue went through the wars having to have a rush hysterectomy in June, after some timely results revealed future risks. -

Guide to the Flora of the Carolinas, Virginia, and Georgia, Working Draft of 17 March 2004 -- LILIACEAE
Guide to the Flora of the Carolinas, Virginia, and Georgia, Working Draft of 17 March 2004 -- LILIACEAE LILIACEAE de Jussieu 1789 (Lily Family) (also see AGAVACEAE, ALLIACEAE, ALSTROEMERIACEAE, AMARYLLIDACEAE, ASPARAGACEAE, COLCHICACEAE, HEMEROCALLIDACEAE, HOSTACEAE, HYACINTHACEAE, HYPOXIDACEAE, MELANTHIACEAE, NARTHECIACEAE, RUSCACEAE, SMILACACEAE, THEMIDACEAE, TOFIELDIACEAE) As here interpreted narrowly, the Liliaceae constitutes about 11 genera and 550 species, of the Northern Hemisphere. There has been much recent investigation and re-interpretation of evidence regarding the upper-level taxonomy of the Liliales, with strong suggestions that the broad Liliaceae recognized by Cronquist (1981) is artificial and polyphyletic. Cronquist (1993) himself concurs, at least to a degree: "we still await a comprehensive reorganization of the lilies into several families more comparable to other recognized families of angiosperms." Dahlgren & Clifford (1982) and Dahlgren, Clifford, & Yeo (1985) synthesized an early phase in the modern revolution of monocot taxonomy. Since then, additional research, especially molecular (Duvall et al. 1993, Chase et al. 1993, Bogler & Simpson 1995, and many others), has strongly validated the general lines (and many details) of Dahlgren's arrangement. The most recent synthesis (Kubitzki 1998a) is followed as the basis for familial and generic taxonomy of the lilies and their relatives (see summary below). References: Angiosperm Phylogeny Group (1998, 2003); Tamura in Kubitzki (1998a). Our “liliaceous” genera (members of orders placed in the Lilianae) are therefore divided as shown below, largely following Kubitzki (1998a) and some more recent molecular analyses. ALISMATALES TOFIELDIACEAE: Pleea, Tofieldia. LILIALES ALSTROEMERIACEAE: Alstroemeria COLCHICACEAE: Colchicum, Uvularia. LILIACEAE: Clintonia, Erythronium, Lilium, Medeola, Prosartes, Streptopus, Tricyrtis, Tulipa. MELANTHIACEAE: Amianthium, Anticlea, Chamaelirium, Helonias, Melanthium, Schoenocaulon, Stenanthium, Veratrum, Toxicoscordion, Trillium, Xerophyllum, Zigadenus. -

Nuytsia the Journal of the Western Australian Herbarium 31: 197–202 Published Online 20 August 2020
T.D. Macfarlane, A.P. Brown & C.J. French, Wurmbea flavanthera (Colchicaceae), a new species 197 Nuytsia The journal of the Western Australian Herbarium 31: 197–202 Published online 20 August 2020 Wurmbea flavanthera (Yellow-anthered Wurmbea, Colchicaceae), a new species from Western Australia’s Mid West region Terry D. Macfarlane1, Andrew P. Brown and Christopher J. French Western Australian Herbarium, Biodiversity and Conservation Science, Department of Biodiversity, Conservation and Attractions, Locked Bag 104, Bentley Delivery Centre, Western Australia 6983 1Corresponding author, email: [email protected] SHORT COMMUNICATION Wurmbea densiflora (Benth.) T.Macfarlane (Colchicaceae) is an attractive, small geophyte bearing several pink flowers, occurring in the northern Wheatbelt and adjacent rangelands of Western Australia. However, the species concept used in a revision of Wurmbea Thunb. in Australia (Macfarlane 1980, 1987) proves to be a mixture of two species, W. densiflora in the original sense of Bentham (1878) and a later-flowering, undescribed species. This second species was given the phrase nameW . sp. Paynes Find (C.J. French 1237), by which it has been known for some time (Western Australian Herbarium 1998–). The new species, having been well-researched in the field and Herbarium, is formally described below. The previous confusion between the two species was partly due to a lack of field knowledge and an inadequate appreciation of the relatively constrained flowering times ofWurmbea species. Wurmbea flavanthera T.Macfarlane, A.P.Br. & C.J.French, sp. nov. Type: 11.8 km north of Jose Street, Mullewa, on Carnarvon – Mullewa Road, west of road on a minor track, Western Australia, 17 August 2011, T.D. -

Partial Flora Survey Rottnest Island Golf Course
PARTIAL FLORA SURVEY ROTTNEST ISLAND GOLF COURSE Prepared by Marion Timms Commencing 1 st Fairway travelling to 2 nd – 11 th left hand side Family Botanical Name Common Name Mimosaceae Acacia rostellifera Summer scented wattle Dasypogonaceae Acanthocarpus preissii Prickle lily Apocynaceae Alyxia Buxifolia Dysentry bush Casuarinacea Casuarina obesa Swamp sheoak Cupressaceae Callitris preissii Rottnest Is. Pine Chenopodiaceae Halosarcia indica supsp. Bidens Chenopodiaceae Sarcocornia blackiana Samphire Chenopodiaceae Threlkeldia diffusa Coast bonefruit Chenopodiaceae Sarcocornia quinqueflora Beaded samphire Chenopodiaceae Suada australis Seablite Chenopodiaceae Atriplex isatidea Coast saltbush Poaceae Sporabolis virginicus Marine couch Myrtaceae Melaleuca lanceolata Rottnest Is. Teatree Pittosporaceae Pittosporum phylliraeoides Weeping pittosporum Poaceae Stipa flavescens Tussock grass 2nd – 11 th Fairway Family Botanical Name Common Name Chenopodiaceae Sarcocornia quinqueflora Beaded samphire Chenopodiaceae Atriplex isatidea Coast saltbush Cyperaceae Gahnia trifida Coast sword sedge Pittosporaceae Pittosporum phyliraeoides Weeping pittosporum Myrtaceae Melaleuca lanceolata Rottnest Is. Teatree Chenopodiaceae Sarcocornia blackiana Samphire Central drainage wetland commencing at Vietnam sign Family Botanical Name Common Name Chenopodiaceae Halosarcia halecnomoides Chenopodiaceae Sarcocornia quinqueflora Beaded samphire Chenopodiaceae Sarcocornia blackiana Samphire Poaceae Sporobolis virginicus Cyperaceae Gahnia Trifida Coast sword sedge -

Alphabetical Lists of the Vascular Plant Families with Their Phylogenetic
Colligo 2 (1) : 3-10 BOTANIQUE Alphabetical lists of the vascular plant families with their phylogenetic classification numbers Listes alphabétiques des familles de plantes vasculaires avec leurs numéros de classement phylogénétique FRÉDÉRIC DANET* *Mairie de Lyon, Espaces verts, Jardin botanique, Herbier, 69205 Lyon cedex 01, France - [email protected] Citation : Danet F., 2019. Alphabetical lists of the vascular plant families with their phylogenetic classification numbers. Colligo, 2(1) : 3- 10. https://perma.cc/2WFD-A2A7 KEY-WORDS Angiosperms family arrangement Summary: This paper provides, for herbarium cura- Gymnosperms Classification tors, the alphabetical lists of the recognized families Pteridophytes APG system in pteridophytes, gymnosperms and angiosperms Ferns PPG system with their phylogenetic classification numbers. Lycophytes phylogeny Herbarium MOTS-CLÉS Angiospermes rangement des familles Résumé : Cet article produit, pour les conservateurs Gymnospermes Classification d’herbier, les listes alphabétiques des familles recon- Ptéridophytes système APG nues pour les ptéridophytes, les gymnospermes et Fougères système PPG les angiospermes avec leurs numéros de classement Lycophytes phylogénie phylogénétique. Herbier Introduction These alphabetical lists have been established for the systems of A.-L de Jussieu, A.-P. de Can- The organization of herbarium collections con- dolle, Bentham & Hooker, etc. that are still used sists in arranging the specimens logically to in the management of historical herbaria find and reclassify them easily in the appro- whose original classification is voluntarily pre- priate storage units. In the vascular plant col- served. lections, commonly used methods are systema- Recent classification systems based on molecu- tic classification, alphabetical classification, or lar phylogenies have developed, and herbaria combinations of both. -

Kuntheria Pedunculata (F.Muell.) Conran & Clifford Family: Colchicaceae Conran, J.G
Australian Tropical Rainforest Plants - Online edition Kuntheria pedunculata (F.Muell.) Conran & Clifford Family: Colchicaceae Conran, J.G. & Clifford, H.T. (1987) Flora of Australia 45: 491. Stem Flowers and fruits as a multistemmed shrub about 1-2 m tall. Leaves Twigs zig-zagged. Leaves +/- distichous with about 5-7 main longitudinal veins including the midrib. Leaf blades about 6-18 x 2-6 cm, petioles about 0.1-0.5 cm long. Reticulate veins run +/- at right angles to the secondary veins like the rungs of a ladder. Petiole grooved or channelled on the upper surface. Flowers Tepals about 7-9 mm long. Stamens about 6-8 mm long, filaments about 3-4 mm long, anthers about 3-4 mm long. Each tepal folded around a stamen in the flower bud but opening to release them as Flowers. CC-BY J.L. Dowe the flower matures. Pollen yellow. Ovary about 3-4 mm diam., ovules numerous in each locule. Style about 1-2 mm long. Fruit Capsule 3-lobed, about 8-15 mm diam., styles persistent at the apex. Seeds white when fresh, about 7-9 x 4-6 mm, irregular in shape. Endosperm hard. Embryo very small, about 0.5 mm long, cylindrical. Seedlings Leaves and flowers. CC-BY J.L. Usually one cataphyll, bract-like, about 4-6 mm long. First pair of true leaves triplinerved. Leaf base Dowe clasping the stem at a point where it is distinctly bent. At the tenth leaf stage: leaf blade with about 5- 7 longitudinal veins on either side of the midrib. -

Tracing History
Comprehensive Summaries of Uppsala Dissertations from the Faculty of Science and Technology 911 Tracing History Phylogenetic, Taxonomic, and Biogeographic Research in the Colchicum Family BY ANNIKA VINNERSTEN ACTA UNIVERSITATIS UPSALIENSIS UPPSALA 2003 Dissertation presented at Uppsala University to be publicly examined in Lindahlsalen, EBC, Uppsala, Friday, December 12, 2003 at 10:00 for the degree of Doctor of Philosophy. The examination will be conducted in English. Abstract Vinnersten, A. 2003. Tracing History. Phylogenetic, Taxonomic and Biogeographic Research in the Colchicum Family. Acta Universitatis Upsaliensis. Comprehensive Summaries of Uppsala Dissertations from the Faculty of Science and Technology 911. 33 pp. Uppsala. ISBN 91-554-5814-9 This thesis concerns the history and the intrafamilial delimitations of the plant family Colchicaceae. A phylogeny of 73 taxa representing all genera of Colchicaceae, except the monotypic Kuntheria, is presented. The molecular analysis based on three plastid regions—the rps16 intron, the atpB- rbcL intergenic spacer, and the trnL-F region—reveal the intrafamilial classification to be in need of revision. The two tribes Iphigenieae and Uvularieae are demonstrated to be paraphyletic. The well-known genus Colchicum is shown to be nested within Androcymbium, Onixotis constitutes a grade between Neodregea and Wurmbea, and Gloriosa is intermixed with species of Littonia. Two new tribes are described, Burchardieae and Tripladenieae, and the two tribes Colchiceae and Uvularieae are emended, leaving four tribes in the family. At generic level new combinations are made in Wurmbea and Gloriosa in order to render them monophyletic. The genus Androcymbium is paraphyletic in relation to Colchicum and the latter genus is therefore expanded. -
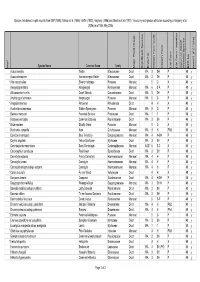
BFS048 Site Species List
Species lists based on plot records from DEP (1996), Gibson et al. (1994), Griffin (1993), Keighery (1996) and Weston et al. (1992). Taxonomy and species attributes according to Keighery et al. (2006) as of 16th May 2005. Species Name Common Name Family Major Plant Group Significant Species Endemic Growth Form Code Growth Form Life Form Life Form - aquatics Common SSCP Wetland Species BFS No kens01 (FCT23a) Wd? Acacia sessilis Wattle Mimosaceae Dicot WA 3 SH P 48 y Acacia stenoptera Narrow-winged Wattle Mimosaceae Dicot WA 3 SH P 48 y * Aira caryophyllea Silvery Hairgrass Poaceae Monocot 5 G A 48 y Alexgeorgea nitens Alexgeorgea Restionaceae Monocot WA 6 S-R P 48 y Allocasuarina humilis Dwarf Sheoak Casuarinaceae Dicot WA 3 SH P 48 y Amphipogon turbinatus Amphipogon Poaceae Monocot WA 5 G P 48 y * Anagallis arvensis Pimpernel Primulaceae Dicot 4 H A 48 y Austrostipa compressa Golden Speargrass Poaceae Monocot WA 5 G P 48 y Banksia menziesii Firewood Banksia Proteaceae Dicot WA 1 T P 48 y Bossiaea eriocarpa Common Bossiaea Papilionaceae Dicot WA 3 SH P 48 y * Briza maxima Blowfly Grass Poaceae Monocot 5 G A 48 y Burchardia congesta Kara Colchicaceae Monocot WA 4 H PAB 48 y Calectasia narragara Blue Tinsel Lily Dasypogonaceae Monocot WA 4 H-SH P 48 y Calytrix angulata Yellow Starflower Myrtaceae Dicot WA 3 SH P 48 y Centrolepis drummondiana Sand Centrolepis Centrolepidaceae Monocot AUST 6 S-C A 48 y Conostephium pendulum Pearlflower Epacridaceae Dicot WA 3 SH P 48 y Conostylis aculeata Prickly Conostylis Haemodoraceae Monocot WA 4 H P 48 y Conostylis juncea Conostylis Haemodoraceae Monocot WA 4 H P 48 y Conostylis setigera subsp. -

Gondwanan Origin of Major Monocot Groups Inferred from Dispersal-Vicariance Analysis Kåre Bremer Uppsala University
Aliso: A Journal of Systematic and Evolutionary Botany Volume 22 | Issue 1 Article 3 2006 Gondwanan Origin of Major Monocot Groups Inferred from Dispersal-Vicariance Analysis Kåre Bremer Uppsala University Thomas Janssen Muséum National d'Histoire Naturelle Follow this and additional works at: http://scholarship.claremont.edu/aliso Part of the Botany Commons Recommended Citation Bremer, Kåre and Janssen, Thomas (2006) "Gondwanan Origin of Major Monocot Groups Inferred from Dispersal-Vicariance Analysis," Aliso: A Journal of Systematic and Evolutionary Botany: Vol. 22: Iss. 1, Article 3. Available at: http://scholarship.claremont.edu/aliso/vol22/iss1/3 Aliso 22, pp. 22-27 © 2006, Rancho Santa Ana Botanic Garden GONDWANAN ORIGIN OF MAJOR MONO COT GROUPS INFERRED FROM DISPERSAL-VICARIANCE ANALYSIS KARE BREMERl.3 AND THOMAS JANSSEN2 lDepartment of Systematic Botany, Evolutionary Biology Centre, Norbyvagen l8D, SE-752 36 Uppsala, Sweden; 2Museum National d'Histoire Naturelle, Departement de Systematique et Evolution, USM 0602: Taxonomie et collections, 16 rue Buffon, 75005 Paris, France 3Corresponding author ([email protected]) ABSTRACT Historical biogeography of major monocot groups was investigated by biogeographical analysis of a dated phylogeny including 79 of the 81 monocot families using the Angiosperm Phylogeny Group II (APG II) classification. Five major areas were used to describe the family distributions: Eurasia, North America, South America, Africa including Madagascar, and Australasia including New Guinea, New Caledonia, and New Zealand. In order to investigate the possible correspondence with continental breakup, the tree with its terminal distributions was fitted to the geological area cladogram «Eurasia, North America), (Africa, (South America, Australasia») and to alternative area cladograms using the TreeFitter program. -

Biodiversity and Ecology of Critically Endangered, Rûens Silcrete Renosterveld in the Buffeljagsrivier Area, Swellendam
Biodiversity and Ecology of Critically Endangered, Rûens Silcrete Renosterveld in the Buffeljagsrivier area, Swellendam by Johannes Philippus Groenewald Thesis presented in fulfilment of the requirements for the degree of Masters in Science in Conservation Ecology in the Faculty of AgriSciences at Stellenbosch University Supervisor: Prof. Michael J. Samways Co-supervisor: Dr. Ruan Veldtman December 2014 Stellenbosch University http://scholar.sun.ac.za Declaration I hereby declare that the work contained in this thesis, for the degree of Master of Science in Conservation Ecology, is my own work that have not been previously published in full or in part at any other University. All work that are not my own, are acknowledge in the thesis. ___________________ Date: ____________ Groenewald J.P. Copyright © 2014 Stellenbosch University All rights reserved ii Stellenbosch University http://scholar.sun.ac.za Acknowledgements Firstly I want to thank my supervisor Prof. M. J. Samways for his guidance and patience through the years and my co-supervisor Dr. R. Veldtman for his help the past few years. This project would not have been possible without the help of Prof. H. Geertsema, who helped me with the identification of the Lepidoptera and other insect caught in the study area. Also want to thank Dr. K. Oberlander for the help with the identification of the Oxalis species found in the study area and Flora Cameron from CREW with the identification of some of the special plants growing in the area. I further express my gratitude to Dr. Odette Curtis from the Overberg Renosterveld Project, who helped with the identification of the rare species found in the study area as well as information about grazing and burning of Renosterveld. -

The Vascular System of Monocotyledonous Stems Author(S): Martin H
The Vascular System of Monocotyledonous Stems Author(s): Martin H. Zimmermann and P. B. Tomlinson Source: Botanical Gazette, Vol. 133, No. 2 (Jun., 1972), pp. 141-155 Published by: The University of Chicago Press Stable URL: http://www.jstor.org/stable/2473813 . Accessed: 30/08/2011 15:50 Your use of the JSTOR archive indicates your acceptance of the Terms & Conditions of Use, available at . http://www.jstor.org/page/info/about/policies/terms.jsp JSTOR is a not-for-profit service that helps scholars, researchers, and students discover, use, and build upon a wide range of content in a trusted digital archive. We use information technology and tools to increase productivity and facilitate new forms of scholarship. For more information about JSTOR, please contact [email protected]. The University of Chicago Press is collaborating with JSTOR to digitize, preserve and extend access to Botanical Gazette. http://www.jstor.org 1972] McCONNELL& STRUCKMEYER ALAR AND BORON-DEFICIENTTAGETES 141 tomato, turnip and cotton to variations in boron nutri- Further investigationson the relation of photoperiodto tion. II. Anatomical responses. BOT.GAZ. 118:53-71. the boron requirementsof plants. BOT.GAZ. 109:237-249. REED, D. J., T. C. MOORE, and J. D. ANDERSON. 1965. Plant WATANABE,R., W. CHORNEY,J. SKOK,and S. H. WENDER growth retardant B-995: a possible mode of action. 1964. Effect of boron deficiency on polyphenol produc- Science 148: 1469-1471. tion in the sunflower.Phytochemistry 3:391-393. SKOK, J. 1957. Relationships of boron nutrition to radio- ZEEVAART,J. A. D. 1966. Inhibition of stem growth and sensitivity of sunflower plants. -

Evolutionary History of Floral Key Innovations in Angiosperms Elisabeth Reyes
Evolutionary history of floral key innovations in angiosperms Elisabeth Reyes To cite this version: Elisabeth Reyes. Evolutionary history of floral key innovations in angiosperms. Botanics. Université Paris Saclay (COmUE), 2016. English. NNT : 2016SACLS489. tel-01443353 HAL Id: tel-01443353 https://tel.archives-ouvertes.fr/tel-01443353 Submitted on 23 Jan 2017 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. NNT : 2016SACLS489 THESE DE DOCTORAT DE L’UNIVERSITE PARIS-SACLAY, préparée à l’Université Paris-Sud ÉCOLE DOCTORALE N° 567 Sciences du Végétal : du Gène à l’Ecosystème Spécialité de Doctorat : Biologie Par Mme Elisabeth Reyes Evolutionary history of floral key innovations in angiosperms Thèse présentée et soutenue à Orsay, le 13 décembre 2016 : Composition du Jury : M. Ronse de Craene, Louis Directeur de recherche aux Jardins Rapporteur Botaniques Royaux d’Édimbourg M. Forest, Félix Directeur de recherche aux Jardins Rapporteur Botaniques Royaux de Kew Mme. Damerval, Catherine Directrice de recherche au Moulon Président du jury M. Lowry, Porter Curateur en chef aux Jardins Examinateur Botaniques du Missouri M. Haevermans, Thomas Maître de conférences au MNHN Examinateur Mme. Nadot, Sophie Professeur à l’Université Paris-Sud Directeur de thèse M.