Genetic Divergence Studies in Indigenous Malus Baccata Biotypes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Department of Planning and Zoning
Department of Planning and Zoning Subject: Howard County Landscape Manual Updates: Recommended Street Tree List (Appendix B) and Recommended Plant List (Appendix C) - Effective July 1, 2010 To: DLD Review Staff Homebuilders Committee From: Kent Sheubrooks, Acting Chief Division of Land Development Date: July 1, 2010 Purpose: The purpose of this policy memorandum is to update the Recommended Plant Lists presently contained in the Landscape Manual. The plant lists were created for the first edition of the Manual in 1993 before information was available about invasive qualities of certain recommended plants contained in those lists (Norway Maple, Bradford Pear, etc.). Additionally, diseases and pests have made some other plants undesirable (Ash, Austrian Pine, etc.). The Howard County General Plan 2000 and subsequent environmental and community planning publications such as the Route 1 and Route 40 Manuals and the Green Neighborhood Design Guidelines have promoted the desirability of using native plants in landscape plantings. Therefore, this policy seeks to update the Recommended Plant Lists by identifying invasive plant species and disease or pest ridden plants for their removal and prohibition from further planting in Howard County and to add other available native plants which have desirable characteristics for street tree or general landscape use for inclusion on the Recommended Plant Lists. Please note that a comprehensive review of the street tree and landscape tree lists were conducted for the purpose of this update, however, only -

Osher Lifelong Learning Institute
USDA-ARS National Plant Germplasm System Conservation of Fruit & Nut Genetic Resources Joseph Postman Plant Pathologist & Curator National Clonal Germplasm Repository Corvallis, Oregon May 2010 Mission: Collect – Preserve Evaluate – Enhance - Distribute World Diversity of Plant Genetic Resources for Improving the Quality and Production of Economic Crops Important to U.S. and World Agriculture Apple Accessions at Geneva Malus angustifolia ( 59 Accessions) Malus sikkimensis ( 14 Accessions) Malus baccata ( 67 Accessions) Malus sp. ( 41 Accessions) Malus bhutanica ( 117 Accessions) Malus spectabilis ( 9 Accessions) Malus brevipes ( 2 Accessions) Malus sylvestris ( 70 Accessions) Malus coronaria ( 98 Accessions) Malus toringo ( 122 Accessions) Malus domestica ( 1,389 Accessions) Malus transitoria ( 63 Accessions) Malus doumeri ( 2 Accessions) Malus trilobata ( 2 Accessions) Malus florentina ( 4 Accessions) Malus tschonoskii ( 3 Accessions) Malus floribunda ( 12 Accessions) Malus x adstringens ( 2 Accessions) Malus fusca ( 147 Accessions) Malus x arnoldiana ( 2 Accessions) Malus halliana ( 15 Accessions) Malus x asiatica ( 20 Accessions) Malus honanensis ( 4 Accessions) Malus x astracanica ( 1 Accessions) Malus hupehensis ( 185 Accessions) Malus x atrosanguinea ( 2 Accessions) Malus hybrid ( 337 Accessions) Malus x dawsoniana ( 2 Accessions) Malus ioensis ( 72 Accessions) Malus x hartwigii ( 5 Accessions) Malus kansuensis ( 45 Accessions) Malus x magdeburgensis ( 2 Accessions) Malus komarovii ( 1 Accessions) Malus x micromalus ( 25 Accessions) -

Maine Coefficient of Conservatism
Coefficient of Coefficient of Scientific Name Common Name Nativity Conservatism Wetness Abies balsamea balsam fir native 3 0 Abies concolor white fir non‐native 0 Abutilon theophrasti velvetleaf non‐native 0 3 Acalypha rhomboidea common threeseed mercury native 2 3 Acer ginnala Amur maple non‐native 0 Acer negundo boxelder non‐native 0 0 Acer pensylvanicum striped maple native 5 3 Acer platanoides Norway maple non‐native 0 5 Acer pseudoplatanus sycamore maple non‐native 0 Acer rubrum red maple native 2 0 Acer saccharinum silver maple native 6 ‐3 Acer saccharum sugar maple native 5 3 Acer spicatum mountain maple native 6 3 Acer x freemanii red maple x silver maple native 2 0 Achillea millefolium common yarrow non‐native 0 3 Achillea millefolium var. borealis common yarrow non‐native 0 3 Achillea millefolium var. millefolium common yarrow non‐native 0 3 Achillea millefolium var. occidentalis common yarrow non‐native 0 3 Achillea ptarmica sneezeweed non‐native 0 3 Acinos arvensis basil thyme non‐native 0 Aconitum napellus Venus' chariot non‐native 0 Acorus americanus sweetflag native 6 ‐5 Acorus calamus calamus native 6 ‐5 Actaea pachypoda white baneberry native 7 5 Actaea racemosa black baneberry non‐native 0 Actaea rubra red baneberry native 7 3 Actinidia arguta tara vine non‐native 0 Adiantum aleuticum Aleutian maidenhair native 9 3 Adiantum pedatum northern maidenhair native 8 3 Adlumia fungosa allegheny vine native 7 Aegopodium podagraria bishop's goutweed non‐native 0 0 Coefficient of Coefficient of Scientific Name Common Name Nativity -

Managing Fire Blight by Choosing Decreased Host Susceptibility Levels and Rootstock Traits , 2020 , January 15 January
Managing Fire Blight by Choosing Decreased Host Susceptibility Levels and Rootstock Traits , 2020 , January 15 January Awais Khan Plant Pathology and Plant-microbe Biology, SIPS, Cornell University, Geneva, NY F ire blight bacterial infection of apple cells Khan et al. 2013 Host resistance and fire blight management in apple orchards Host resistance is considered most sustainable option for disease management due to Easy to deploy/implement in the orchards Low input and cost-effective Environment friendly No choice to the growers--most of the new and old cultivars are highly susceptible Apple breeding to develop resistant cultivars Domestication history of the cultivated apple 45-50 Malus species-----Malus sieversii—Gene flow Malus baccata Diameter: 1 cm Malus sieversii Malus baccata Diameter: up to 8 cm Malus orientalis Diameter: 2-4 cm Malus sylvestris Diameter: 1-3 cm Duan et al. 2017 Known sources of major/moderate resistance to fire blight to breed resistant cultivars Source Resistance level Malus Robusta 5 80% Malus Fusca 66% Malus Arnoldiana, Evereste, Malus floribunda 821 35-55% Fiesta, Enterprise 34-46% • Fruit quality is the main driver for success of an apple cultivar • Due to long juvenility of apples, it can take 20-25 years to breed resistance from wild crab apples Genetic disease resistance in world’s largest collection of apples Evaluation of fire blight resistance of accessions from US national apple collection o Grafted 5 replications: acquired bud-wood and rootstocks o Inoculated with Ea273 Erwinia amylovora strain -

Finding List and Guide to Tllc Secrest Arboretum
SPECIAL CIRCULAR 91 MAY 1960 REV ISED Finding List and Guide to tllc Secrest Arboretum Wood Utilization Research Laboratory and Secrest Arboretum Headquarters Effective July 1, 1965, the name of the Ohio Agricultural Experiment Station was changed to: 0 Ohio Agricu ltura I Research and Development Center, ')ll Wooster, Ohio • This page intentionally blank. • GUIBE TO THE SECREST ARBORETUM OHIO AGRICULTURAL EXPERJMENT STATION WOOSTER, OHIO '/ By John E. Aughanbaugh, Harry R. Muckley, and Oliver D. Diller* In May 1950 the forest and ornamental plantings at the Ohio Agricultural Experiment Station were dedicated as the Secrest Arboretum in memory of Edmund Secrest, the father of forestry in Ohio. Since 19o8 these plantings have been expanded to include well over 600 species and varieties of trees and shrubs from many parts of the world. It is the purpose of this publication to serve as a finding list and guide to the Arboretum. The Purpose of the Arboretum One purpose of the Arboretum is to _determine the species and varieties of trees adapted for ornamental, windbreak, and shelterbelt uses in 6hio. There are many varieties of spruces, firs, yews, arborvitae, and other coniferous trees and shrubs growing here for observation by people interested in landscaping and the planting of shelterbelts. Among the more recent additions to the Arboretum is a collection of over 60 varieties of flowering crabapples, 57 selections of hollies and a plot of Chinese dawnredwood. The second purpose of the Arboretum is to determine the spe?ies of trees best adapted for reforestation in Ohio and to determine the silvicultural requirements that will obtain best results in growth and maturity. -

Biodiversity in Karnali Province: Current Status and Conservation
Biodiversity in Karnali Province: Current Status and Conservation Karnali Province Government Ministry of Industry, Tourism, Forest and Environment Surkhet, Nepal Biodiversity in Karnali Province: Current Status and Conservation Karnali Province Government Ministry of Industry, Tourism, Forest and Environment Surkhet, Nepal Copyright: © 2020 Ministry of Industry, Tourism, Forest and Environment, Karnali Province Government, Surkhet, Nepal The views expressed in this publication do not necessarily reflect those of Ministry of Tourism, Forest and Environment, Karnali Province Government, Surkhet, Nepal Editors: Krishna Prasad Acharya, PhD and Prakash K. Paudel, PhD Technical Team: Achyut Tiwari, PhD, Jiban Poudel, PhD, Kiran Thapa Magar, Yogendra Poudel, Sher Bahadur Shrestha, Rajendra Basukala, Sher Bahadur Rokaya, Himalaya Saud, Niraj Shrestha, Tejendra Rawal Production Editors: Prakash Basnet and Anju Chaudhary Reproduction of this publication for educational or other non-commercial purposes is authorized without prior written permission from the copyright holder provided the source is fully acknowledged. Reproduction of this publication for resale or other commercial purposes is prohibited without prior written permission of the copyright holder. Citation: Acharya, K. P., Paudel, P. K. (2020). Biodiversity in Karnali Province: Current Status and Conservation. Ministry of Industry, Tourism, Forest and Environment, Karnali Province Government, Surkhet, Nepal Cover photograph: Tibetan wild ass in Limi valley © Tashi R. Ghale Keywords: biodiversity, conservation, Karnali province, people-wildlife nexus, biodiversity profile Editors’ Note Gyau Khola Valley, Upper Humla © Geraldine Werhahn This book “Biodiversity in Karnali Province: Current Status and Conservation”, is prepared to consolidate existing knowledge about the state of biodiversity in Karnali province. The book presents interrelated dynamics of society, physical environment, flora and fauna that have implications for biodiversity conservation. -

Melatonin Improves Waterlogging Tolerance of Malus Baccata (Linn.) Borkh
fpls-08-00483 April 3, 2017 Time: 15:7 # 1 ORIGINAL RESEARCH published: 05 April 2017 doi: 10.3389/fpls.2017.00483 Melatonin Improves Waterlogging Tolerance of Malus baccata (Linn.) Borkh. Seedlings by Maintaining Aerobic Respiration, Photosynthesis and ROS Migration Xiaodong Zheng1, Jingzhe Zhou2, Dun-Xian Tan3, Na Wang1, Lin Wang1, Dongqian Shan1 and Jin Kong1* 1 College of Horticulture, China Agricultural University, Beijing, China, 2 Beijing Soil and Fertilizer Work Station, Beijing, China, 3 Department of Cellular and Structural Biology, UT Health Science Center San Antonio, San Antonio, TX, USA Waterlogging, one of the notorious abiotic stressors, retards the growth of apple plants and reduces their production. Thus, it is an urgent agenda for scientists to identify the suitable remedies for this problem. In the current study, we found that melatonin significantly improved the tolerance of apple seedlings against waterlogging stress. This was indicated by the reduced chlorosis and wilting of the seedlings after melatonin applications either by leaf spray or root irrigation. The mechanisms involve Edited by: in that melatonin functions to maintain aerobic respiration, preserves photosynthesis Wei Hu, and reduces oxidative damage of the plants which are under waterlogging stress. Institute of Tropical Bioscience and Biotechnology, Chinese Academy of Melatonin application also enhances the gene expression of its synthetic enzymes Tropical Agricultural Sciences, China (MbT5H1, MbAANAT3, MbASMT9) and increases melatonin production. This is the Reviewed by: first report of a positive feedback that exogenous melatonin application promotes the Shi Xiao, Sun Yat-sen University, China melatonin synthesis in plants. A post-transcriptional regulation apparently participated Wei Fan, in this regulation. -

Sonicator Dyeing of Cotton, Wool and Silk with the Leaves Extract
Volume 6, Issue 1, Spring2009 Sonicator Dyeing of Cotton, Wool and Silk with the Leaves Extract Padma S Vankar*a, Rakhi Shanker a, Shalini Dixit a and Debajit Mahantab aFacility for Ecological and Analytical Testing Indian Institute of Technology, Kanpur-208 016, India bArunachal Pradesh State Council for Science & Technology Vivek-Vihar, Itanagar-791 113, Arunachal Pradesh ABSTRACT Malus sikkimensis. (Local name- Chap shaw sheng) belongs to family Rosaceae is being primarily used for preparing tea by Monpas tribes of Arunachal Pradesh. In the present study sonicator dyeing with Malus extract has been demonstrated. Pretreatment with 1-2 % metal mordant and using 5 % of plant extract (owf) was found to be optimum and showed very good hue colors for cotton, wool and silk dyed fabrics. We have demonstrated that the use of sonication in conjunction with metal mordanting has a synergistic effect on better dye uptake, dye adherence and eventually on good fastness properties of the dyed swatches of cotton, silk and wool. Keywords: Malus sikkimensis, natural dye, cotton, wool, silk, commercial dyeing 1. Introduction: Malus sikkimensis, belongs to family Rosaceae, is an evergreen woody tree with a maximum height of 40 m and diameter 2.3 m. It occurs in humid valley forest of Arunachal Pradesh. In the present study Malus leaves (figure-1) have been used as natural dye-stuff, which is primarily used as a substitute for tea by the local Arunachalees. The crude aqueous extract has been shown to dye cotton, silk and wool samples as shown in figures 5-7. Figure-1 Leaves of Malus sikkimensis Traditional Knowledge of its use: Stem and leaves of the plant locally known as 2. -
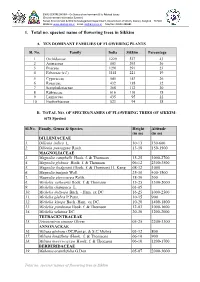
1. Total No. Species/ Name of Flowering Trees in Sikkim
ENVIS CENTRE SIKKIM – On Status of environment & Its Related Issues (Environmental Information System) Forest, Environment & Wildlife Management Department, Government of Sikkim, Deorali, Gangtok – 737102 Website: www.sikenvis.nic.in Email: [email protected] Tele/Fax: 03592-280381 1. Total no. species/ name of flowering trees in Sikkim A. TEN DOMINANT FAMILIES OF FLOWERING PLANTS Sl. No. Family India Sikkim Percentage 1 Orchidaceae 1229 527 43 2 Asteraceae 803 293 36 3 Poaceae 1291 291 23 4 Fabaceae (s.l.) 1141 221 19 5 Cyperaceae 545 143 26 6 Rosaceae 432 138 32 7 Scrophulariaceae 368 112 30 8 Rubiaceae 616 110 18 9 Lamiaceae 435 95 22 10 Euphorbiaceae 523 94 18 B. TOTAL NO. OF SPECIES/NAMES OF FLOWERING TREES OF SIKKIM: (678 Species) Sl.No. Family, Genus & Species Height Altitude (in m) (in m) DILLENIACEAE 1. Dillenia indica L. 10-13 150-600 2. Dillenia pentagyna Roxb. 13-18 150-1500 MAGNOLIACEAE 3. Magnolia campbelli Hook. f. & Thomson 15-25 1000-2700 4. Magnolia globosa Hook. f. & Thomson 06-12 2300-3500 5. Magnolia hodgsonii (Hook. f. & Thomson) H. Keng 08-12 1000 6. Magnolia insignis Wall. 25-30 400-1500 7. Magnolia pterocarpa Roxb. 18-30 500 8. Michelia cathcartii Hook. f. & Thomson 15-25 1500-2000 9. Michelia champaca L. 03-05 10. Michelia doltsopa Buch. - Ham. ex DC. 16-25 1000-2500 11. Michelia glabra P.Pann. 10-15 900 12. Michelia kisopa Buch.-Ham. ex DC. 10-20 1400-1800 13. Michelia punduana Hook. f. & Thomson 12-03 1000-1600 14. Michelia velutina DC. -

A 33-Year Evaluation of Resistance and Pathogenicity in the Apple
HORTSCIENCE 44(3):599–608. 2009. tification of scab-resistant cultivars is impor- tant for both commercial producers and retailers of crabapples (Romer et al., 2003) A 33-year Evaluation of Resistance and for commercial apple breeding programs that seek to develop scab-resistant apples and Pathogenicity in the Apple (Janick, 2002; Shay et al., 1962). Host plant resistance is considered one of Scab–crabapples Pathosystem the most efficient and effective methods to control plant diseases with much of the Janna Beckerman1 breeding effort within the genus Malus Department of Botany and Plant Pathology, Purdue University, 915 directed to evaluate resistance to apple scab. West State Street, West Lafayette, IN 47907 The V. inaequalis–Malus interaction is one of the earliest examples demonstrating gene- James Chatfield for-gene interactions (Boone, 1971; Hough, Department of Horticulture and Crop Science, The Ohio Agricultural 1944; Williams and Shay, 1957). The gene- for-gene theory states that for every major Research and Development Center, Wooster, OH 44691-4096 gene conferring resistance (R) in the host, Erik Draper there exists a corresponding avirulence (AVR) gene in the pathogen (Flor, 1956; Hammond- The Ohio State University Extension Service, Geauga County, OH Kosack and Jones, 1997). Disease resistance 44021-9521 results only when the corresponding product Additional index words. Malus, Venturia inaequalis, avirulence, durable resistance of a dominant resistance gene (R) recognizes a dominant Avr gene product from the path- Abstract. Crabapples (Malus spp.) are popular ornamental trees in the commercial and ogen. Disease results when the loss or alter- residential landscape. Over a 33-year period at the Secrest Arboretum, Wooster, OH, 287 ation of the pathogen avirulence gene (now accessions of ornamental crabapple were evaluated for their resistance to apple scab denoted as avr) fails to trigger recognition by caused by the fungus Venturia inaequalis. -

Title: Genetic Variation of Crabapples ( Malus Spp.) Found on Governors Island and NYC Area
Title: Genetic Variation of Crabapples ( Malus spp.) found on Governors Island and NYC Area Team Members: Jianri Chen, Zinan Ma, Iulius Sergiu Moldovan and Xuanzhi Zhao Sponsoring Teacher: Alfred Lwin School: Manhattan Comprehensive Night & Day High School Crab Apple Crabapple variation? YEAH IT’S CRABAPPLE!!!!! Medicinal uses “Accelerate cerebral circulation and oxygenation and regulates the secretion of neuromodulators to help facilitate intellectual work To treat constipation and worms and mental fatigue” http://www.forresthealth.com/Crab-Apple-Bud. html “Crabapple buds are rich in beneficial phyto-chemicals such growth factors and plant hormones, enzymes, nucleic acids, oligoelements, and phytonutrients such as polyphenols and flavonoids.” http://www.goldenneedleonline.com/Crab-Apple- Bud.html Crabapple for jelly and cider Crabapple fruits have sour and bitter taste if you eat them raw. But the fruits can be made jelly and cider and crabapple wood can also be used to grill and smoke food to add aroma Abstracts Crabapple trees are one of the most common flowering fruit trees found along streets and parks and around landmark buildings in NYC. Their colorful blossom and different size fruits make them one of the most popular ornament trees for landscape. In fact, apples and crabapples belong to the rose family, Rosacea and genus Malus. All over the world there are approximately 35 different species of crabapple and 700 cultivars and they can be hybrids that result from more than one species. Our observation of the crab apple samples showed some variations in physical characteristic and therefore we hypothesized that those crabapple trees found on Governors Island, NY and in other NYC area may have come from those 35 species or hybrids of 700 cultivars and there may be genetic variation among them. -

The Vulnerability of US Apple (Malus) Genetic Resources
Genet Resour Crop Evol (2015) 62:765–794 DOI 10.1007/s10722-014-0194-2 RESEARCH ARTICLE The vulnerability of US apple (Malus) genetic resources Gayle M. Volk • C. Thomas Chao • Jay Norelli • Susan K. Brown • Gennaro Fazio • Cameron Peace • Jim McFerson • Gan-Yuan Zhong • Peter Bretting Received: 20 June 2014 / Accepted: 27 October 2014 / Published online: 13 November 2014 Ó Springer Science+Business Media Dordrecht (outside the USA) 2014 Abstract Apple (Malus 9 domestica Borkh.) is one wide range of biotic and abiotic stress resistances as of the top three US fruit crops in production and value. well as desirable productivity and fruit quality attri- Apple production has high costs for land, labor and butes. However, access to wild materials is limited and inputs, and orchards are a long-term commitment. wild Malus throughout the world is at risk of loss due Production is dominated by only a few apple scion and to human encroachment and changing climatic pat- rootstock cultivars, which increases its susceptibility terns. The USDA-ARS National Plant Germplasm to dynamic external threats. Apple crop wild relatives, System (NPGS) Malus collection, maintained by the including progenitor species Malus sieversii (Ledeb.) Plant Genetic Resources Unit in Geneva, NY, US is M. Roem., Malus orientalis Uglitzk., Malus sylvestris among the largest collections of cultivated apple and (L.) Mill., and Malus prunifolia (Willd.) Borkh., as Malus species in the world. The collection currently well as many other readily hybridized species, have a has 5004 unique accessions in the field and 1603 seed accessions representing M. 9 domestica,33Malus species, and 15 hybrid species.