1 Development of Prediction Models of Methane Production by Sheep
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Review Article: Inhibition of Methanogenic Archaea by Statins As a Targeted Management Strategy for Constipation and Related Disorders
Alimentary Pharmacology and Therapeutics Review article: inhibition of methanogenic archaea by statins as a targeted management strategy for constipation and related disorders K. Gottlieb*, V. Wacher*, J. Sliman* & M. Pimentel† *Synthetic Biologics, Inc., Rockville, SUMMARY MD, USA. † Gastroenterology, Cedars-Sinai Background Medical Center, Los Angeles, CA, USA. Observational studies show a strong association between delayed intestinal transit and the production of methane. Experimental data suggest a direct inhibitory activity of methane on the colonic and ileal smooth muscle and Correspondence to: a possible role for methane as a gasotransmitter. Archaea are the only con- Dr K. Gottlieb, Synthetic Biologics, fi Inc., 9605 Medical Center Drive, rmed biological sources of methane in nature and Methanobrevibacter Rockville, MD 20850, USA. smithii is the predominant methanogen in the human intestine. E-mail: [email protected] Aim To review the biosynthesis and composition of archaeal cell membranes, Publication data archaeal methanogenesis and the mechanism of action of statins in this context. Submitted 8 September 2015 First decision 29 September 2015 Methods Resubmitted 7 October 2015 Narrative review of the literature. Resubmitted 20 October 2015 Accepted 20 October 2015 Results EV Pub Online 11 November 2015 Statins can inhibit archaeal cell membrane biosynthesis without affecting This uncommissioned review article was bacterial numbers as demonstrated in livestock and humans. This opens subject to full peer-review. the possibility of a therapeutic intervention that targets a specific aetiologi- cal factor of constipation while protecting the intestinal microbiome. While it is generally believed that statins inhibit methane production via their effect on cell membrane biosynthesis, mediated by inhibition of the HMG- CoA reductase, there is accumulating evidence for an alternative or addi- tional mechanism of action where statins inhibit methanogenesis directly. -

Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline
Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline • Phlogenetics of Archaea • Phlogenetics of archaeal lipids • Papers Phyla • Two? main phyla – Euryarchaeota • Methanogens • Extreme halophiles • Extreme thermophiles • Sulfate-reducing – Crenarchaeota • Extreme thermophiles – Korarchaeota? • Hyperthermophiles • indicated only by environmental DNA sequences – Nanoarchaeum? • N. equitans a fast evolving euryarchaeal lineage, not novel, early diverging archaeal phylum – Ancient archael group? • In deepest brances of Crenarchaea? Euryarchaea? Archaeal Lipids • Methanogens – Di- and tetra-ethers of glycerol and isoprenoid alcohols – Core mostly archaeol or caldarchaeol – Core sometimes sn-2- or Images removed due to sn-3-hydroxyarchaeol or copyright considerations. macrocyclic archaeol –PMI • Halophiles – Similar to methanogens – Exclusively synthesize bacterioruberin • Marine Crenarchaea Depositional Archaeal Lipids Biological Origin Environment Crocetane methanotrophs? methane seeps? methanogens, PMI (2,6,10,15,19-pentamethylicosane) methanotrophs hypersaline, anoxic Squalane hypersaline? C31-C40 head-to-head isoprenoids Smit & Mushegian • “Lost” enzymes of MVA pathway must exist – Phosphomevalonate kinase (PMK) – Diphosphomevalonate decarboxylase – Isopentenyl diphosphate isomerase (IPPI) Kaneda et al. 2001 Rohdich et al. 2001 Boucher et al. • Isoprenoid biosynthesis of archaea evolved through a combination of processes – Co-option of ancestral enzymes – Modification of enzymatic specificity – Orthologous and non-orthologous gene -

Characteristics and Metabolic Patterns of Soil Methanogenic Archaea Communities in the High Latitude Natural Wetlands of China
Characteristics and Metabolic Patterns of Soil Methanogenic Archaea Communities in the High Latitude Natural Wetlands of China Di Wu Northeast Forestry University Caihong Zhao Northeast Forestry University Hui Bai Forestry Science Research Institute of Heilongjiang Province Fujuan Feng Northeast Forestry University Xin Sui Heilongjiang University Guangyu Sun ( [email protected] ) Northeast Forestry University Research article Keywords: Wetlands, Methanogens, Community diversity, Indicator species, Methanogenic metabolic patterns Posted Date: August 12th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-54821/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/18 Abstract Background: Soil methanogenic microorganisms are one of the primary methane-producing microbes in wetlands. However, we still poorly understand the community characteristic and metabolic patterns of these microorganisms according to vegetation type and seasonal changes. Therefore, to better elucidate the effects of the vegetation type and seasonal factors on the methanogenic community structure and metabolic patterns, we detected the characteristics of the soil methanogenic mcrA gene from three types of natural wetlands in different seasons in the Xiaoxing'an Mountain region, China. Result: The results indicated that the distribution of Methanobacteriaceae (hydrogenotrophic methanogens) was higher in winter, while Methanosarcinaceae and Methanosaetaceae accounted for a higher proportion in summer. Hydrogenotrophic methanogenesis was the dominant trophic pattern in each wetland. The results of principal coordinate analysis and cluster analysis showed that the vegetation type considerably inuenced the methanogenic community composition. The methanogenic community structure in the Betula platyphylla – Larix gmelinii wetland was relatively different from the structure of the other two wetland types. -
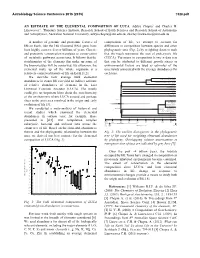
An Estimate of the Elemental Composition of Luca
Astrobiology Science Conference 2015 (2015) 7328.pdf AN ESTIMATE OF THE ELEMENTAL COMPOSITION OF LUCA. Aditya Chopra1 and Charles H. Lineweaver1, 1Planetary Science Institute, Research School of Earth Sciences and Research School of Astronomy and Astrophysics, Australian National University, [email protected], [email protected] A number of genomic and proteomic features of composition of life, we attempt to account for life on Earth, like the 16S ribosomal RNA gene, have differences in composition between species and other been highly conserved over billions of years. Genetic phylogenetic taxa (Fig. 2) by weighting datasets such and proteomic conservation translates to conservation that the result represents the root of prokaryotic life of metabolic pathways across taxa. It follows that the (LUCA). Variations in composition between data sets stoichiometry of the elements that make up some of that can be attributed to different growth stages or the biomolecules will be conserved. By extension, the environmental factors are used as estimates of the elemental make up of the whole organism is a uncertainty associated with the average abundances for relatively conserved feature of life on Earth [1,2]. each taxa. We describe how average bulk elemental Euryarchaeota 1a Methanococci, Methanobacteria, Methanopyri 7 Euryarchaeota 1b abundances in extant life can yield an indirect estimate 6 Thermoplasmata, Methanomicrobia, Halobacteria, Archaeoglobi Euryarchaeota 2 4 Thermococci of relative abundances of elements in the Last Crenarchaeota Sulfolobus, Thermoproteus ? Thaumarchaeota Universal Common Ancestor (LUCA). The results Cenarchaeum ? Korarchaeota ARMAN could give us important hints about the stoichiometry ? 2 Archaeal Richmond Mine Acidophilic Nanoorganisms Nanoarchaeota of the environment where LUCA existed and perhaps Archaea Terrabacteria clues to the processes involved in the origin and early Actinobacteria, Deinococcus-Thermus, 1 Cyanobacteria, Life Chloroflexi, 9 evolution of life [3]. -

Establishment and Analysis of Microbial Communities Capable of Producing Methane from Grass Waste at Extremely High C/N Ratio
International Journal of New Technology and Research (IJNTR) ISSN:2454-4116, Volume-2, Issue-9, September 2016 Pages 81-86 Establishment and Analysis of Microbial Communities Capable of Producing Methane from Grass Waste at Extremely High C/N Ratio S. Matsuda, T. Ohtsuki* Anaerobic digestion is a conventional method for biomass Abstract— Acclimation of microbial communities, aiming to utilization [8]. Despite the fact that a great deal of research methane production from grass as a sole substrate at extremely for methane production from grass has been conducted high carbon-to-nitrogen (C/N) ratio, was conducted. In a series especially in recent years, almost processes need supply of of experiments with various sizes of added grass, two microbial communities showing high methane production were obtained abundant sludge source such as sewage and manure [9, 10]. with powdered grass. In the two microbial communities Many studies indicated that the optimal carbon-to-nitrogen designated NR and RP, Bacteroidia including genera (C/N) ratio in methane fermentation were 25-30, and the C/N Bacteroides, Dysgonomonas, Proteiniphilum, and Alistipes were ratio higher than 40 are not generally suitable [11]. The C/N detected as dominant members in eubacteria. It was also shown ration of raw grass shows a wide range; wild grassy weed is at that Methanomicrobia and Methanobacteria including genera relatively high (>59) while lawn grass is at low (<29) [12]. Methanomassiliicoccus and Methanobacterium were found as dominant members in methanogen. It is noteworthy that Therefore, it is difficult to conduct methane fermentation nitrogen fixation were observed both in NR and RP, suggesting with wild grass-only, also being due to less degradability of that insufficiency of nitrogen sources would be complemented lignocellulose [13]. -

Lifestyle Preferences Drive the Structure and Diversity of Bacterial and Archaeal Communities in a Small Riverine Reservoir
www.nature.com/scientificreports OPEN Lifestyle preferences drive the structure and diversity of bacterial and archaeal communities in a small riverine reservoir Carles Borrego1,2, Sergi Sabater1,3* & Lorenzo Proia1,4 Spatial heterogeneity along river networks is interrupted by dams, afecting the transport, processing, and storage of organic matter, as well as the distribution of biota. We here investigated the structure of planktonic (free-living, FL), particle-attached (PA) and sediment-associated (SD) bacterial and archaeal communities within a small reservoir. We combined targeted-amplicon sequencing of bacterial and archaeal 16S rRNA genes in the DNA and RNA community fractions from FL, PA and SD, followed by imputed functional metagenomics, in order to unveil diferences in their potential metabolic capabilities within the reservoir (tail, mid, and dam sections) and lifestyles (FL, PA, SD). Both bacterial and archaeal communities were structured according to their life-style preferences rather than to their location in the reservoir. Bacterial communities were richer and more diverse when attached to particles or inhabiting the sediment, while Archaea showed an opposing trend. Diferences between PA and FL bacterial communities were consistent at functional level, the PA community showing higher potential capacity to degrade complex carbohydrates, aromatic compounds, and proteinaceous materials. Our results stressed that particle-attached prokaryotes were phylogenetically and metabolically distinct from their free-living counterparts, and that performed as hotspots for organic matter processing within the small reservoir. Spatial heterogeneity of river networks results from the sequence of lotic segments—which promote the transport and quick transformation of materials—and lentic segments such as large pools and wetlands—which mostly contribute to the process and storage of organic matter 1,2. -
![Downloaded from the NCBI FTP Site [37]](https://docslib.b-cdn.net/cover/4020/downloaded-from-the-ncbi-ftp-site-37-1134020.webp)
Downloaded from the NCBI FTP Site [37]
Life 2015, 5, 818-840; doi:10.3390/life5010818 OPEN ACCESS life ISSN 2075-1729 www.mdpi.com/journal/life Article Archaeal Clusters of Orthologous Genes (arCOGs): An Update and Application for Analysis of Shared Features between Thermococcales, Methanococcales, and Methanobacteriales Kira S. Makarova *, Yuri I. Wolf and Eugene V. Koonin National Center for Biotechnology Information, NLM, National Institutes of Health, Bethesda, MD 20894, USA; E-Mails: [email protected] (Y.I.W.); [email protected] (E.V.K.) * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +1-301-435-5913; Fax: +1-301-435-7793. Academic Editors: Hans-Peter Klenk, Michael W. W. Adams and Roger A. Garrett Received: 12 January 2015 / Accepted: 28 February 2015 / Published: 10 March 2015 Abstract: With the continuously accelerating genome sequencing from diverse groups of archaea and bacteria, accurate identification of gene orthology and availability of readily expandable clusters of orthologous genes are essential for the functional annotation of new genomes. We report an update of the collection of archaeal Clusters of Orthologous Genes (arCOGs) to cover, on average, 91% of the protein-coding genes in 168 archaeal genomes. The new arCOGs were constructed using refined algorithms for orthology identification combined with extensive manual curation, including incorporation of the results of several completed and ongoing research projects in archaeal genomics. A new level of classification is introduced, superclusters that unit two or more arCOGs and more completely reflect gene family evolution than individual, disconnected arCOGs. Assessment of the current archaeal genome annotation in public databases indicates that consistent use of arCOGs can significantly improve the annotation quality. -

Salinity Drives Archaeal Distribution Patterns in High Altitude Lake Sediments on the Tibetan Plateau Yongqin Liu 1,2,∗, John C
FEMS Microbiology Ecology, 92, 2016, fiw033 doi: 10.1093/femsec/fiw033 Advance Access Publication Date: 16 February 2016 Research Article RESEARCH ARTICLE Salinity drives archaeal distribution patterns in high altitude lake sediments on the Tibetan Plateau Yongqin Liu 1,2,∗, John C. Priscu3, Jinbo Xiong4, Ralf Conrad5, Trista Vick-Majors3, Haiyan Chu6 and Juzhi Hou1 Downloaded from 1Key Laboratory of Tibetan Environment Changes and Land Surface Processes, Institute of Tibetan Plateau Research, Beijing 100101, China, 2CAS Center for Excellence in Tibetan Plateau Earth Sciences, Chinese Academy of Sciences, Beijing 100101, China, 3Department of Land Resources and Environmental Sciences, Montana State University, Bozeman, MT 59717, USA, 4Faculty of Marine Sciences, Ningbo University, Ningbo http://femsec.oxfordjournals.org/ 315211, China, 5Max Planck Institute for Terrestrial Microbiology, Marburg 35043, Germany and 6State Key Laboratory of Soil and Sustainable Agriculture, Institute of Soil Science, Chinese Academy of Sciences, Nanjing 210008, China ∗Corresponding author: Institute of Tibetan Plateau, Chinese Academy of Science, No. 16, Lincuo Rd., Beijing 100101, China. Tel: 86-10-84097122; E-mail: [email protected] One sentence summary: We represent a comprehensive investigation of archaeal diversity in the pristine lake sediments across the Tibetan Plateau, and found salinity is the key factor controlling archaeal community diversity and composition. Editor: Dirk Wagner by guest on April 5, 2016 ABSTRACT Archaeal communities and the factors regulating their diversity in high altitude lakes are poorly understood. Here, we provide the first high-throughput sequencing study of Archaea from Tibetan Plateau lake sediments. We analyzed twenty lake sediments from the world’s highest and largest plateau and found diverse archaeal assemblages that clustered into groups dominated by methanogenic Euryarchaeota, Crenarchaeota and Halobacteria/mixed euryarchaeal phylotypes. -

Carbon Sources of Benthic Fauna in Temperate Lakes Across Multiple Trophic States
Biogeosciences, 18, 4369–4388, 2021 https://doi.org/10.5194/bg-18-4369-2021 © Author(s) 2021. This work is distributed under the Creative Commons Attribution 4.0 License. Carbon sources of benthic fauna in temperate lakes across multiple trophic states Annika Fiskal1, Eva Anthamatten1, Longhui Deng1, Xingguo Han1, Lorenzo Lagostina1, Anja Michel1, Rong Zhu1, Nathalie Dubois2,3, Carsten J. Schubert1,4, Stefano M. Bernasconi3, and Mark A. Lever1 1Institute of Biogeochemistry and Pollutant Dynamics (IBP), ETH Zurich, Universitätstrasse 16, 8092 Zurich, Switzerland 2Department Surface Waters – Research and Management, Eawag, Swiss Federal Institute of Aquatic Science and Technology, Überlandstrasse 133, 8600 Dübendorf, Switzerland 3Department of Earth Sciences, ETH Zurich, Sonneggstrasse 5, 8092 Zurich, Switzerland 4Department of Surface Waters – Research and Management, Swiss Federal Institute of Aquatic Science and Technology (EAWAG), Seestrasse 79, 6047 Kastanienbaum, Switzerland Correspondence: Annika Fiskal (annikafi[email protected]) and Mark A. Lever ([email protected]) Received: 26 August 2020 – Discussion started: 11 September 2020 Revised: 2 June 2021 – Accepted: 21 June 2021 – Published: 28 July 2021 Abstract. Previous studies have shown that microbially pro- in so far uncharacterized relationships with detrital organic- duced methane can be a dominant carbon source of lacus- matter-degrading bacterial endosymbionts. trine sedimentary macrofauna in eutrophic lakes, most likely through grazing on methane-oxidizing bacteria. Here we in- vestigate the contributions of different carbon sources to macrofaunal biomass across five lakes in central Switzerland 1 Introduction that range from oligotrophic to highly eutrophic. Macrofau- nal communities change with trophic state, with chironomid Lake sediments are globally important organic C sinks (Ein- larvae dominating oligotrophic and tubificid oligochaetes sele et al., 2001; Mendonça et al., 2017) and sources of dominating eutrophic lake sediments. -

Diversity of Methane-Cycling Microorganisms in Soils and Their Relation to Oxygen
Diversity of Methane-cycling Microorganisms in Soils and Their Relation to Oxygen Claudia Knief* Institute of Crop Science and Resource Conservation – Molecular Biology of the Rhizosphere, University of Bonn, Bonn, Germany. *Correspondence: [email protected] htps://doi.org/10.21775/cimb.033.023 Abstract Introduction Microorganisms are important players in the Methane cycling microorganisms are of interest global methane cycle. Anaerobic methanogenic for microbiologists since more than a century. archaea are largely responsible for methane pro- Research on these microorganisms was initially duction, while aerobic methanotrophic bacteria, largely driven by the curiosity to understand their as well as anaerobic methanotrophic bacteria particular physiology that leads to the production and archaea, are involved in methane oxidation. or consumption of methane. While this interest is In anoxic wetland soils, methanogens produce still a driver, the importance of methane as green- methane, while methanotrophs act as a flter house gas has become another important factor, and reduce methane emissions. In the predomi- promoting further research on methanogenic and nantly oxic upland soils, aerobic methanotrophs methanotrophic microorganisms. Tis leads to a oxidize atmospheric methane. Tis review gives continuously beter understanding of their physi- an overview of the diversity of methanogenic ology and ecology, and it becomes evident that and methanotrophic microorganisms, highlights the processes of microbial methane production recent discoveries and provides information and consumption are mediated by more com- concerning their occurrence in soils. Recent plex functional guilds than initially thought. Te fndings indicate that the methanogenic and improved understanding is not only due to the con- methanotrophic lifestyles are more widespread stantly increasing diversity of methanogenic and in microorganisms than previously thought, and methanotrophic microorganisms (e.g. -

Author's Tracked Changes
Carbon sources of benthic fauna in temperate lakes across multiple trophic states Annika Fiskal1, Eva Anthamatten1, Longhui Deng1, Xingguo Han1, Lorenzo Lagostina1, Anja Michel1, Rong Zhu1, Nathalie 5 Dubois2,3, Carsten J. Schubert1,4, Stefano M. Bernasconi3, Mark A. Lever1 1Institute of Biogeochemistry and Pollutant Dynamics (IBP), ETH Zurich, Universitätstrasse 16, 8092 Zurich, Switzerland 2Surface Waters Research – Management, Eawag, Swiss Federal Institute of Aquatic Science and Technology, Überlandstrasse 133, 8600 Dübendorf, Switzerland 10 3Department of Earth Sciences, ETH Zurich, Sonneggstrasse 5, 8092 Zurich, Switzerland 4Department of Surface Waters – Research and Management, Swiss Federal Institute of Aquatic Science and Technology (EAWAG), Seestrasse 79, 6047 Kastanienbaum, Switzerland Correspondence: Annika Fiskal ([email protected]) and Mark A. Lever ([email protected]) 15 Keywords methane-derived carbon, benthic macrofauna, lacustrine sediment, oligochaetes, chironomid larvae, methane-oxidizing bacteria, endosymbionts Abstract. Previous studies have shown that microbially produced methane can be a dominant carbon source of lacustrine 20 sedimentary macrofauna in eutrophic lakes, most likely through grazing on methane-oxidizing bacteria. Here we investigate the contributions of different carbon sources to macrofaunal biomass across five lakes in central Switzerland that range from oligotrophic to highly eutrophic. Macrofaunal communities change with trophic state, with chironomid larvae dominating oligotrophic and tubificid oligochaetes dominating eutrophic lake sediments. 13C-isotopic data suggest that the average contribution of methane-derived carbon to the biomass of both macrofaunal groups is similar, but consistently remains minor, 25 ranging from only ~1% in the oligotrophic lake to at most 12% in the eutrophic lakes. The remaining biomass can be explained with assimilation of detritus-derived organic carbon. -

Diversity, Ecology and Evolution of Archaea
REVIEW ARTICLE https://doi.org/10.1038/s41564-020-0715-z Diversity, ecology and evolution of Archaea Brett J. Baker 1 ✉ , Valerie De Anda1, Kiley W. Seitz1, Nina Dombrowski 1, Alyson E. Santoro2 and Karen G. Lloyd 3 Compared to bacteria, our knowledge of archaeal biology is limited. Historically, microbiologists have mostly relied on cultur- ing and single-gene diversity surveys to understand Archaea in nature. However, only six of the 27 currently proposed archaeal phyla have cultured representatives. Advances in genomic sequencing and computational approaches are revolutionizing our understanding of Archaea. The recovery of genomes belonging to uncultured groups from the environment has resulted in the description of several new phyla, many of which are globally distributed and are among the predominant organisms on the planet. In this Review, we discuss how these genomes, together with long-term enrichment studies and elegant in situ measurements, are providing insights into the metabolic capabilities of the Archaea. We also debate how such studies reveal how important Archaea are in mediating an array of ecological processes, including global carbon and nutrient cycles, and how this increase in archaeal diversity has expanded our view of the tree of life and early archaeal evolution, and has provided new insights into the origin of eukaryotes. nitially, all single-celled, non-eukaryotic microorganisms were 16S rRNA gene sequencing revealed many deeply branching classified as the ‘Prokaryota’1 and, later, the ‘Monera’2. These groups19. This uncultivated diversity is commonly referred to as Iearly classifications lumped Bacteria and Archaea into a single ‘microbial dark matter’. group based primarily on morphology.