Archaea in and on the Human Body: Health Implications and Future Directions
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Methanothermus Fervidus Type Strain (V24S)
UC Davis UC Davis Previously Published Works Title Complete genome sequence of Methanothermus fervidus type strain (V24S). Permalink https://escholarship.org/uc/item/9367m39j Journal Standards in genomic sciences, 3(3) ISSN 1944-3277 Authors Anderson, Iain Djao, Olivier Duplex Ngatchou Misra, Monica et al. Publication Date 2010-11-20 DOI 10.4056/sigs.1283367 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Standards in Genomic Sciences (2010) 3:315-324 DOI:10.4056/sigs.1283367 Complete genome sequence of Methanothermus fervidus type strain (V24ST) Iain Anderson1, Olivier Duplex Ngatchou Djao2, Monica Misra1,3, Olga Chertkov1,3, Matt Nolan1, Susan Lucas1, Alla Lapidus1, Tijana Glavina Del Rio1, Hope Tice1, Jan-Fang Cheng1, Roxanne Tapia1,3, Cliff Han1,3, Lynne Goodwin1,3, Sam Pitluck1, Konstantinos Liolios1, Natalia Ivanova1, Konstantinos Mavromatis1, Natalia Mikhailova1, Amrita Pati1, Evelyne Brambilla4, Amy Chen5, Krishna Palaniappan5, Miriam Land1,6, Loren Hauser1,6, Yun-Juan Chang1,6, Cynthia D. Jeffries1,6, Johannes Sikorski4, Stefan Spring4, Manfred Rohde2, Konrad Eichinger7, Harald Huber7, Reinhard Wirth7, Markus Göker4, John C. Detter1, Tanja Woyke1, James Bristow1, Jonathan A. Eisen1,8, Victor Markowitz5, Philip Hugenholtz1, Hans-Peter Klenk4, and Nikos C. Kyrpides1* 1 DOE Joint Genome Institute, Walnut Creek, California, USA 2 HZI – Helmholtz Centre for Infection Research, Braunschweig, Germany 3 Los Alamos National Laboratory, Bioscience Division, Los Alamos, New Mexico, USA 4 DSMZ - German Collection of Microorganisms and Cell Cultures GmbH, Braunschweig, Germany 5 Biological Data Management and Technology Center, Lawrence Berkeley National Laboratory, Berkeley, California, USA 6 Oak Ridge National Laboratory, Oak Ridge, Tennessee, USA 7 University of Regensburg, Archaeenzentrum, Regensburg, Germany 8 University of California Davis Genome Center, Davis, California, USA *Corresponding author: Nikos C. -

Review Article: Inhibition of Methanogenic Archaea by Statins As a Targeted Management Strategy for Constipation and Related Disorders
Alimentary Pharmacology and Therapeutics Review article: inhibition of methanogenic archaea by statins as a targeted management strategy for constipation and related disorders K. Gottlieb*, V. Wacher*, J. Sliman* & M. Pimentel† *Synthetic Biologics, Inc., Rockville, SUMMARY MD, USA. † Gastroenterology, Cedars-Sinai Background Medical Center, Los Angeles, CA, USA. Observational studies show a strong association between delayed intestinal transit and the production of methane. Experimental data suggest a direct inhibitory activity of methane on the colonic and ileal smooth muscle and Correspondence to: a possible role for methane as a gasotransmitter. Archaea are the only con- Dr K. Gottlieb, Synthetic Biologics, fi Inc., 9605 Medical Center Drive, rmed biological sources of methane in nature and Methanobrevibacter Rockville, MD 20850, USA. smithii is the predominant methanogen in the human intestine. E-mail: [email protected] Aim To review the biosynthesis and composition of archaeal cell membranes, Publication data archaeal methanogenesis and the mechanism of action of statins in this context. Submitted 8 September 2015 First decision 29 September 2015 Methods Resubmitted 7 October 2015 Narrative review of the literature. Resubmitted 20 October 2015 Accepted 20 October 2015 Results EV Pub Online 11 November 2015 Statins can inhibit archaeal cell membrane biosynthesis without affecting This uncommissioned review article was bacterial numbers as demonstrated in livestock and humans. This opens subject to full peer-review. the possibility of a therapeutic intervention that targets a specific aetiologi- cal factor of constipation while protecting the intestinal microbiome. While it is generally believed that statins inhibit methane production via their effect on cell membrane biosynthesis, mediated by inhibition of the HMG- CoA reductase, there is accumulating evidence for an alternative or addi- tional mechanism of action where statins inhibit methanogenesis directly. -

Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline
Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline • Phlogenetics of Archaea • Phlogenetics of archaeal lipids • Papers Phyla • Two? main phyla – Euryarchaeota • Methanogens • Extreme halophiles • Extreme thermophiles • Sulfate-reducing – Crenarchaeota • Extreme thermophiles – Korarchaeota? • Hyperthermophiles • indicated only by environmental DNA sequences – Nanoarchaeum? • N. equitans a fast evolving euryarchaeal lineage, not novel, early diverging archaeal phylum – Ancient archael group? • In deepest brances of Crenarchaea? Euryarchaea? Archaeal Lipids • Methanogens – Di- and tetra-ethers of glycerol and isoprenoid alcohols – Core mostly archaeol or caldarchaeol – Core sometimes sn-2- or Images removed due to sn-3-hydroxyarchaeol or copyright considerations. macrocyclic archaeol –PMI • Halophiles – Similar to methanogens – Exclusively synthesize bacterioruberin • Marine Crenarchaea Depositional Archaeal Lipids Biological Origin Environment Crocetane methanotrophs? methane seeps? methanogens, PMI (2,6,10,15,19-pentamethylicosane) methanotrophs hypersaline, anoxic Squalane hypersaline? C31-C40 head-to-head isoprenoids Smit & Mushegian • “Lost” enzymes of MVA pathway must exist – Phosphomevalonate kinase (PMK) – Diphosphomevalonate decarboxylase – Isopentenyl diphosphate isomerase (IPPI) Kaneda et al. 2001 Rohdich et al. 2001 Boucher et al. • Isoprenoid biosynthesis of archaea evolved through a combination of processes – Co-option of ancestral enzymes – Modification of enzymatic specificity – Orthologous and non-orthologous gene -

Histone Variants in Archaea and the Evolution of Combinatorial Chromatin Complexity
Histone variants in archaea and the evolution of combinatorial chromatin complexity Kathryn M. Stevensa,b, Jacob B. Swadlinga,b, Antoine Hochera,b, Corinna Bangc,d, Simonetta Gribaldoe, Ruth A. Schmitzc, and Tobias Warneckea,b,1 aMolecular Systems Group, Quantitative Biology Section, Medical Research Council London Institute of Medical Sciences, London W12 0NN, United Kingdom; bInstitute of Clinical Sciences, Faculty of Medicine, Imperial College London, London W12 0NN, United Kingdom; cInstitute for General Microbiology, University of Kiel, 24118 Kiel, Germany; dInstitute of Clinical Molecular Biology, University of Kiel, 24105 Kiel, Germany; and eDepartment of Microbiology, Unit “Evolutionary Biology of the Microbial Cell,” Institut Pasteur, 75015 Paris, France Edited by W. Ford Doolittle, Dalhousie University, Halifax, NS, Canada, and approved October 28, 2020 (received for review April 14, 2020) Nucleosomes in eukaryotes act as platforms for the dynamic inte- additional histone dimers can be taggedontothistetramertoyield gration of epigenetic information. Posttranslational modifications oligomers of increasing length that wrap correspondingly more DNA are reversibly added or removed and core histones exchanged for (3, 6–9). Almost all archaeal histones lack tails and PTMs have yet to paralogous variants, in concert with changing demands on tran- be reported. Many archaea do, however, encode multiple histone scription and genome accessibility. Histones are also common in paralogs (8, 10) that can flexibly homo- and heterodimerize in -

Characteristics and Metabolic Patterns of Soil Methanogenic Archaea Communities in the High Latitude Natural Wetlands of China
Characteristics and Metabolic Patterns of Soil Methanogenic Archaea Communities in the High Latitude Natural Wetlands of China Di Wu Northeast Forestry University Caihong Zhao Northeast Forestry University Hui Bai Forestry Science Research Institute of Heilongjiang Province Fujuan Feng Northeast Forestry University Xin Sui Heilongjiang University Guangyu Sun ( [email protected] ) Northeast Forestry University Research article Keywords: Wetlands, Methanogens, Community diversity, Indicator species, Methanogenic metabolic patterns Posted Date: August 12th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-54821/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/18 Abstract Background: Soil methanogenic microorganisms are one of the primary methane-producing microbes in wetlands. However, we still poorly understand the community characteristic and metabolic patterns of these microorganisms according to vegetation type and seasonal changes. Therefore, to better elucidate the effects of the vegetation type and seasonal factors on the methanogenic community structure and metabolic patterns, we detected the characteristics of the soil methanogenic mcrA gene from three types of natural wetlands in different seasons in the Xiaoxing'an Mountain region, China. Result: The results indicated that the distribution of Methanobacteriaceae (hydrogenotrophic methanogens) was higher in winter, while Methanosarcinaceae and Methanosaetaceae accounted for a higher proportion in summer. Hydrogenotrophic methanogenesis was the dominant trophic pattern in each wetland. The results of principal coordinate analysis and cluster analysis showed that the vegetation type considerably inuenced the methanogenic community composition. The methanogenic community structure in the Betula platyphylla – Larix gmelinii wetland was relatively different from the structure of the other two wetland types. -

Root Microbiota Assembly and Adaptive Differentiation Among European
bioRxiv preprint doi: https://doi.org/10.1101/640623; this version posted May 17, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Root microbiota assembly and adaptive differentiation among European 2 Arabidopsis populations 3 4 Thorsten Thiergart1,7, Paloma Durán1,7, Thomas Ellis2, Ruben Garrido-Oter1,3, Eric Kemen4, Fabrice 5 Roux5, Carlos Alonso-Blanco6, Jon Ågren2,*, Paul Schulze-Lefert1,3,*, Stéphane Hacquard1,*. 6 7 1Max Planck Institute for Plant Breeding Research, 50829 Cologne, Germany 8 2Department of Ecology and Genetics, Evolutionary Biology Centre, Uppsala University, SE‐752 36 9 Uppsala, Sweden 10 3Cluster of Excellence on Plant Sciences (CEPLAS), Max Planck Institute for Plant Breeding Research, 11 50829 Cologne, Germany 12 4Department of Microbial Interactions, IMIT/ZMBP, University of Tübingen, 72076 Tübingen, 13 Germany 14 5LIPM, INRA, CNRS, Université de Toulouse, 31326 Castanet-Tolosan, France 15 6Departamento de Genética Molecular de Plantas, Centro Nacional de Biotecnología (CNB), Consejo 16 Superior de Investigaciones Científicas (CSIC), 28049 Madrid, Spain 17 7These authors contributed equally: Thorsten Thiergart, Paloma Durán 18 *e-mail: [email protected], [email protected], [email protected] 19 20 Summary 21 Factors that drive continental-scale variation in root microbiota and plant adaptation are poorly 22 understood. We monitored root-associated microbial communities in Arabidopsis thaliana and co- 23 occurring grasses at 17 European sites across three years. -
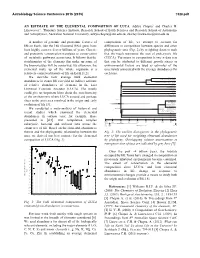
An Estimate of the Elemental Composition of Luca
Astrobiology Science Conference 2015 (2015) 7328.pdf AN ESTIMATE OF THE ELEMENTAL COMPOSITION OF LUCA. Aditya Chopra1 and Charles H. Lineweaver1, 1Planetary Science Institute, Research School of Earth Sciences and Research School of Astronomy and Astrophysics, Australian National University, [email protected], [email protected] A number of genomic and proteomic features of composition of life, we attempt to account for life on Earth, like the 16S ribosomal RNA gene, have differences in composition between species and other been highly conserved over billions of years. Genetic phylogenetic taxa (Fig. 2) by weighting datasets such and proteomic conservation translates to conservation that the result represents the root of prokaryotic life of metabolic pathways across taxa. It follows that the (LUCA). Variations in composition between data sets stoichiometry of the elements that make up some of that can be attributed to different growth stages or the biomolecules will be conserved. By extension, the environmental factors are used as estimates of the elemental make up of the whole organism is a uncertainty associated with the average abundances for relatively conserved feature of life on Earth [1,2]. each taxa. We describe how average bulk elemental Euryarchaeota 1a Methanococci, Methanobacteria, Methanopyri 7 Euryarchaeota 1b abundances in extant life can yield an indirect estimate 6 Thermoplasmata, Methanomicrobia, Halobacteria, Archaeoglobi Euryarchaeota 2 4 Thermococci of relative abundances of elements in the Last Crenarchaeota Sulfolobus, Thermoproteus ? Thaumarchaeota Universal Common Ancestor (LUCA). The results Cenarchaeum ? Korarchaeota ARMAN could give us important hints about the stoichiometry ? 2 Archaeal Richmond Mine Acidophilic Nanoorganisms Nanoarchaeota of the environment where LUCA existed and perhaps Archaea Terrabacteria clues to the processes involved in the origin and early Actinobacteria, Deinococcus-Thermus, 1 Cyanobacteria, Life Chloroflexi, 9 evolution of life [3]. -

Establishment and Analysis of Microbial Communities Capable of Producing Methane from Grass Waste at Extremely High C/N Ratio
International Journal of New Technology and Research (IJNTR) ISSN:2454-4116, Volume-2, Issue-9, September 2016 Pages 81-86 Establishment and Analysis of Microbial Communities Capable of Producing Methane from Grass Waste at Extremely High C/N Ratio S. Matsuda, T. Ohtsuki* Anaerobic digestion is a conventional method for biomass Abstract— Acclimation of microbial communities, aiming to utilization [8]. Despite the fact that a great deal of research methane production from grass as a sole substrate at extremely for methane production from grass has been conducted high carbon-to-nitrogen (C/N) ratio, was conducted. In a series especially in recent years, almost processes need supply of of experiments with various sizes of added grass, two microbial communities showing high methane production were obtained abundant sludge source such as sewage and manure [9, 10]. with powdered grass. In the two microbial communities Many studies indicated that the optimal carbon-to-nitrogen designated NR and RP, Bacteroidia including genera (C/N) ratio in methane fermentation were 25-30, and the C/N Bacteroides, Dysgonomonas, Proteiniphilum, and Alistipes were ratio higher than 40 are not generally suitable [11]. The C/N detected as dominant members in eubacteria. It was also shown ration of raw grass shows a wide range; wild grassy weed is at that Methanomicrobia and Methanobacteria including genera relatively high (>59) while lawn grass is at low (<29) [12]. Methanomassiliicoccus and Methanobacterium were found as dominant members in methanogen. It is noteworthy that Therefore, it is difficult to conduct methane fermentation nitrogen fixation were observed both in NR and RP, suggesting with wild grass-only, also being due to less degradability of that insufficiency of nitrogen sources would be complemented lignocellulose [13]. -

How Your Body Decides If Bacteria Are Friends Or Foes § Would You
§How your body decides if bacteria are friends or foes § Would you: • Let you child eat food that dropped on the ground? • Let your child suck their thumbs? • Take antibiotics without knowing the true reason you are feeling sick? Humans and Microbes • Leeuwenhoek’s discovery of microorganisms in 17th century led people to suspect they might cause diseases • Robert Koch (1876) offered proof of what is now considered germ theory of disease; showed Bacillus anthracis causes anthrax • Today, we now know that most of the bacteria we associate with are not pathogens, and many are critical for our health. Bacteria Are Ubiquitous § We contact numerous microorganisms daily • Every surface on earth is covered! • Even clouds have microbes – Could play a role in seeding rain.. • Some have tremendous commercial value • Yogurts, wine, cheese, vinegar, pickles, etc. • Our bodies: • Breathe in, ingest, pick up on skin • Vast majority do not make us sick, or cause infections • Some colonize body surfaces; or slough off with dead epithelial cells • Most that are swallowed die in stomach or are eliminated in feces • Relatively few are pathogens that cause damage Microbes, Health, and Disease § Most microbes are harmless • Many are beneficial • Normal microbiota (normal flora) are organisms that routinely reside on body’s surfaces • Relationship is a balance, and some can cause disease under certain conditions-- opportunistic infections • Weaknesses in innate or adaptive defenses can leave individuals vulnerable to invasion – malnutrition, cancer, AIDS or -

1 Development of Prediction Models of Methane Production by Sheep
Development of Prediction Models of Methane Production by Sheep and Cows Using Rumen Microbiota Data Thesis Presented in Partial Fulfillment of the Requirements for the Degree Master of Science in the Graduate School of The Ohio State University By Boyang Zhang Graduate Program in Animal Sciences The Ohio State University 2018 Master's Examination Committee: Dr. Zhongtang Yu, Advisor Dr. Moraes, Co-advisor Dr. Firkins 1 Copyrighted by Boyang Zhang 2018 2 Abstract Methane emission from the rumen leads to approximately 10 % loss of the ingested energy, at the conversion of digestible to metabolizable energy. Besides research on mitigation of methane emission, much research interests have also been gravitated towards development of prediction models of methane emissions from livestock because the global warming is reducing agriculture productivity (Johnson and Johnson, 1995). An accurate prediction of enteric methane production from cattle and sheep can assist in balancing the increased livestock production with subsequent environmental impacts. Methane is an inevitable byproduct of the microbial fermentation processes in the rumen, and certain ruminal microbes have direct impacts to methane production (Morgavi et al., 2010). Thus, we hypothesized that the inclusion of individual microbial groups as predictor variables could improve the robustness and accuracy of prediction models. However, inclusion of microbial variables into prediction models can result in overfitting. Machine-learning algorithms can automatically select the key explanatory predictors, and Linear Mixed Models can provide a framework to predict random effects describing between animal variations. We proposed three novel frameworks for subset selections of microbial variables (MV) and one framework for generalized linear mixed models (GLMM) using L1-penalization (GLMMLASSO) selection with cross-validations (CV) ii to address the overfitting problems and to develop parsimonious prediction models of methane production. -

Lifestyle Preferences Drive the Structure and Diversity of Bacterial and Archaeal Communities in a Small Riverine Reservoir
www.nature.com/scientificreports OPEN Lifestyle preferences drive the structure and diversity of bacterial and archaeal communities in a small riverine reservoir Carles Borrego1,2, Sergi Sabater1,3* & Lorenzo Proia1,4 Spatial heterogeneity along river networks is interrupted by dams, afecting the transport, processing, and storage of organic matter, as well as the distribution of biota. We here investigated the structure of planktonic (free-living, FL), particle-attached (PA) and sediment-associated (SD) bacterial and archaeal communities within a small reservoir. We combined targeted-amplicon sequencing of bacterial and archaeal 16S rRNA genes in the DNA and RNA community fractions from FL, PA and SD, followed by imputed functional metagenomics, in order to unveil diferences in their potential metabolic capabilities within the reservoir (tail, mid, and dam sections) and lifestyles (FL, PA, SD). Both bacterial and archaeal communities were structured according to their life-style preferences rather than to their location in the reservoir. Bacterial communities were richer and more diverse when attached to particles or inhabiting the sediment, while Archaea showed an opposing trend. Diferences between PA and FL bacterial communities were consistent at functional level, the PA community showing higher potential capacity to degrade complex carbohydrates, aromatic compounds, and proteinaceous materials. Our results stressed that particle-attached prokaryotes were phylogenetically and metabolically distinct from their free-living counterparts, and that performed as hotspots for organic matter processing within the small reservoir. Spatial heterogeneity of river networks results from the sequence of lotic segments—which promote the transport and quick transformation of materials—and lentic segments such as large pools and wetlands—which mostly contribute to the process and storage of organic matter 1,2. -

The Neuroprotective Effect of Tea Polyphenols on the Regulation of Intestinal Flora
molecules Review The Neuroprotective Effect of Tea Polyphenols on the Regulation of Intestinal Flora Zhicheng Zhang 1,2,3, Yuting Zhang 4, Junmin Li 2,3,*, Chengxin Fu 1,* and Xin Zhang 4,* 1 Key Laboratory of Conservation Biology for Endangered Wildlife of the Ministry of Education, College of Life Sciences, Zhejiang University, Hangzhou 310058, China; [email protected] 2 Taizhou Biomedical Industry Research Institute Co., Ltd., Taizhou 317000, China 3 College of Life Sciences, Taizhou University, Taizhou 317000, China 4 Department of Food Science and Engineering, Ningbo University, Ningbo 315211, China; [email protected] * Correspondence: [email protected] (J.L.); [email protected] (C.F.); [email protected] (X.Z.) Abstract: Tea polyphenols (TPs) are the general compounds of natural polyhydroxyphenols extracted in tea. Although a large number of studies have shown that TPs have obvious neuroprotective and neuro repair effects, they are limited due to the low bioavailability in vivo. However, TPs can act indirectly on the central nervous system by affecting the “microflora–gut–brain axis”, in which the microbiota and its composition represent a factor that determines brain health. Bidirectional communication between the intestinal microflora and the brain (microbe–gut–brain axis) occurs through a variety of pathways, including the vagus nerve, immune system, neuroendocrine pathways, and bacteria-derived metabolites. This axis has been shown to influence neurotransmission and behavior, which is usually associated with neuropsychiatric disorders. In this review, we discuss that TPs and their metabolites may provide benefits by restoring the imbalance of intestinal microbiota and Citation: Zhang, Z.; Zhang, Y.; Li, J.; that TPs are metabolized by intestinal flora, to provide a new idea for TPs to play a neuroprotective Fu, C.; Zhang, X.