Lidar As a Tool for Remote Sensing of Moose Forage Biomass
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Conservation Status of Birds of Prey and Owls in Norway
Conservation status of birds of prey and owls in Norway Oddvar Heggøy & Ingar Jostein Øien Norsk Ornitologisk Forening 2014 NOF-BirdLife Norway – Report 1-2014 © NOF-BirdLife Norway E-mail: [email protected] Publication type: Digital document (pdf)/75 printed copies January 2014 Front cover: Boreal owl at breeding site in Nord-Trøndelag. © Ingar Jostein Øien Editor: Ingar Jostein Øien Recommended citation: Heggøy, O. & Øien, I. J. (2014) Conservation status of birds of prey and owls in Norway. NOF/BirdLife Norway - Report 1-2014. 129 pp. ISSN: 0805-4932 ISBN: 978-82-78-52092-5 Some amendments and addenda have been made to this PDF document compared to the 75 printed copies: Page 25: Picture of snowy owl and photo caption added Page 27: Picture of white-tailed eagle and photo caption added Page 36: Picture of eagle owl and photo caption added Page 58: Table 4 - hen harrier - “Total population” corrected from 26-147 pairs to 26-137 pairs Page 60: Table 5 - northern goshawk –“Total population” corrected from 1434 – 2036 pairs to 1405 – 2036 pairs Page 80: Table 8 - Eurasian hobby - “Total population” corrected from 119-190 pairs to 142-190 pairs Page 85: Table 10 - peregrine falcon – Population estimate for Hedmark corrected from 6-7 pairs to 12-13 pairs and “Total population” corrected from 700-1017 pairs to 707-1023 pairs Page 78: Photo caption changed Page 87: Last paragraph under “Relevant studies” added. Table text increased NOF-BirdLife Norway – Report 1-2014 NOF-BirdLife Norway – Report 1-2014 SUMMARY Many of the migratory birds of prey species in the African-Eurasian region have undergone rapid long-term declines in recent years. -

Gea Norvegica Geopark NORWAY
Gea Norvegica Geopark NORWAY … the never ending story of landscapes United Nations Gea Norvegica Educational, Scientific and UNESCO Cultural Organization Global Geopark Front cover photo: MØLEN. Larvik Photo by Johannes Fredriksen What is Gea Norvegica Geopark? Gea Norvegica Geopark is located in southeastern Norway. You can visit the seaside; beautiful coastal landscapes with lots of islands, inlets and low-relief fiords. Or visit the inland, where mountainous landscapes are raising more than 800 m above sea level in the northern part of the Geopark. Between these two different areas, you will find rich agricultural land, small towns, river valleys and forests. These diverse landscapes are also reflecting the unique geological history of Gea Norvegica Geopark; a story 1500 million years long, about impressive old mountain chains, today worn down, a large tropical sea with old forms of life, rifting continents, NORWAY ancient volcanoes and several ice ages. But it is also the story of how man has used the geological resources, from Stone Age to present. The beautiful and varied nature of southern Norway offers lots of activities, like hiking To learn more about us visit: www.geoparken.no in the mountains, fishing, sailing at the coast and paddling along rivers and lakes. Or you can go shopping, visit local museums or one of the climbing parks. Our local Geopark partners are active in cultivating local culture and traditions. Oslo n Through guided tours or on your own, you can experience the unique landscapes of the Geopark. In addition, through leaflets, maps and digital information, you can get a deeper understanding of how the landscapes was formed through this long geological history and how society has developed based upon resources like soils, limestone, iron and other elements. -

Møte Med Kommunestyrene I TNT Kommunereformen
KOMMUNEREFORM OG EMBETSSTRUKTUR Forvaltningsrammer for tjenesteutvikling Fagdag for helse og omsorgstjenester til psykisk utviklingshemmede Fagdirektør Petter Lodden, Fylkesmannen i Vestfold NB! Spør og kom gjerne med synspunkter underveis! DAGENS TEMA • Kommunereformens hvorfor • Hva er en kommune? • Hva betyr kommunestrukturen? • Hva er viktig for lokaldemokratiet? • Hvorfor er enkelte Vestfold-kommuner for små? • Hva skjer med kommunestrukturen i Vestfold? • Men først litt om Fylkesmannen • Og til sist litt om Fylkesmannens struktur framover Hvorfor kommunereform? • Gode, likeverdige og rettssikre tjenester, uansett bosted: • Tilstrekkelig kapasitet og relevant kompetanse • Tilstrekkelig distanse • Effektive tjenester og administrasjon • Helhetlig og samordnet samfunnsutvikling • «rydde opp» i delte byer og lokalsamfunn • Bærekraftige kommuner, faglig og økonomisk • Styrket lokaldemokrati og rammestyring • Mindre overstyring (stat og fylkeskommune) • Mindre avhengighet av interkommunalt samarbeid 1969-2014: Niårig grunnskole, beredskap mot forurensing , bosetting av flyktninger, avfallshåndtering og avløp, kommuneleger, helsestasjon, somatiske sykehjem, psykisk utviklingshemmede, miljøvern, undervisning barn i institusjoner, vilt- og naturforvaltning, land- brukskontorene, lovfestet rett til 10-årig grunn-skole, SFO, musikk- og kulturskoler, LAR, psykisk helse, fastlegeordning, forhandlingsansvar for lærere, klinisk veterinærvakt, lovfestet rett til barnehage- plass, krisesentre, kvalifiseringsprogrammet, 1964: nasjonalparkstyrer, -

Se Også Våre Hjemmesider: Side 6 Side 5
NR. 2 - 2019 Side 5 Side 6 Tårnagenthelg Koret Lova i Larvik kirke Side 12 Side 10 Konfirmanter på Sverigetur Kirkevalget 2019 • Å R G A N G 7 3 Se også våre hjemmesider: www.larvik.kirken.no • Forsidebilder av kirkene: Bjørnar Berge KONTAKTINFORMASJON Hedrum menighet: Leder i menighetsrådet: Kjell Østby mobil 982 31 266 Kirketjener/klokker: Astrid Helene Floen mobil 938 44 037 Kateket: Lasse Rasmussen Moskvil mobil 467 98 879 Kvelde menighet: Leder i menighetsrådet: Vibeke Kvalevåg mobil 456 04 526 Kirketjener/klokker: Morten Meidell-Pritzier Hagelund mobil 913 81 815 Hvarnes menighet: Leder i menighetsrådet: Geir Bårnes mobil 952 81 991 Kirketjener/klokker: Marit Thorvaldsen mobil 920 99 703 Lardal menighet: Leder i menighetsrådet: Inger Lene Røsholt mobil 977 03 971 Kirketjener i Hem kirke og leder for kirkegårdsdelen i Lardal: Jørn Olav Lie mobil 916 27 537 Kirketjener i Svarstad kirke: Kai Berntsen mobil 932 49 183 Kirketjener i Styrvoll kirke: Annette Gran mobil 910 07 987 Menighetspedagog/diakoniarbeider: Mariann Eskedal mobil 971 99 722 IKT-ansvarlig: Lasse Moskvil mobil 467 98 879 Diakoniarbeider/teamleder: Ove Henning Berrefjord mobil 922 63 088 Kantor i Hedrum, Kvelde og Hvarnes: Jørn Odén Evertsen mobil 936 05 519 Kantor i Hem, Svarstad og Styrvold: Kjell Håkestad mobil 988 51 800 Ønsker du å snakke med en av prestene våre? Eller kanskje du vet om noen som ønsker et besøk? Ta gjerne kontakt: Sokneprest i Kvelde og Hvarnes: Randi Abelsen Woldseth på mobil 414 66 928, e-post: [email protected] Sokneprest i Hedrum: Thorir Jökull Thorsteinsson på mobil 416 42 328, e-post: [email protected] Sokneprest i Lardal: Thorild Simonsen Svanemyr på mobil 412 36 922, e-post: [email protected] Menighetskontor for Hedrum, Kvelde og Hvarnes menigheter sammen med Larvik prosti og Larvik kirkelige fellesråd: Romberggata 4, 3256 Larvik, Postboks 258 Sentrum, 3251 Larvik. -

Kommunereformen I Vestfold - Fylkesmannens Oppsummering Og Tilrådning
Kommunal- og moderniseringsdepartementet Postboks 8112 Dep 0032 OSLO Vår saksbehandler / telefon: Deres referanse: Vår referanse: Vår dato: Fred-Ivar Syrstad 2014/4267 30.06.2016 33 37 11 56 Arkivnr: 310 Kommunereformen i Vestfold - Fylkesmannens oppsummering og tilrådning Vi viser til brev av 30. november 2015 og 7. juni 2016 fra Kommunal- og moderniseringsdepartementet. 1. Fylkesmannens tilrådning om kommunestruktur i Vestfold Frivillige sammenslåingsvedtak fører til at 14 Vestfold-kommuner blir til 6. Den nye kommunestrukturen i fylket vil i all hovedsak tilfredsstille regjeringens og Stortingets mål med kommunereformen. Vestfold-kommunene har selv tegnet det nye kommunekartet. Alle landets kommuner ble høsten 2014 invitert til å delta i prosesser med sikte på å vurdere og avklare om det er aktuelt å slå seg sammen med nabokommuner. Stortingsflertallet oppfordret kommunene til selv å finne frem til sammenslåingsløsninger. Ved behandlingen av kommuneproposisjonen for 2016 fremholdt stortingsflertallet at «alle kommunar har utredningsplikt i kommunereforma. ... Dette inneber blant anna å ...ta stilling til om og i så fall kva kommunar ein ønskjer å gå saman med.» I Vestfold fulgte kommunene oppfordringen fra regjeringen og Stortinget. Kommunene i Vestfold har selv valgt hvem de vil danne nye kommuner sammen med. Uten unntak er det de små kommunene som har tatt initiativet overfor større nabokommuner. Gjennom kongelige resolusjoner, basert på kommunenes egne vedtak, blir Stokke, Andebu og Sandefjord slått sammen fra 2017, Larvik og Lardal fra 2018, Hof og Holmestrand fra 2018 og Nøtterøy og Tjøme fra 2018. Svelvik har fattet vedtak om at kommunen ønsker sammenslåing med Drammen i Buskerud og eventuelt andre kommuner i Drammensregionen fra 2020. -

Kommunesamarbeidet I Vestfold
W Kommunesamarbeidet i Vestfold VIKS. Selskapsinformasjon 2015 Selskapsnavn Vestfold Interkommunale Kontrollutvalgssekretariat - VIKS Organisasjonsform §27 Organisasjonsnummer 987 665 483 Etableringsdato 04.11. 2004 Formål Utøve sekretariatfunksjon for deltakernes kontrollutvalg. Selskapet kan også utføre oppdrag for andre kommuner/fylkeskommuner. Eiere og eierandeler Eiere Horten kommune Holmestrand kommune (“k eierandel) Tønsberg kommune Larvikkommune Sande kommune Vestfold fvlkeskommune Andebu kommune Lardal kommune Nøtterøy kommune Re kommune Hof kommune Tjøme kommune Styresammensetning Verv Navn Honorar kr Styreleder Jan Nærsnes (Horten) 20.000 +1.000 pr. møte Styrenestleder Harald Haug Andersen(Tønsberg) 1.000 pr.møte Medlem Tone Helgesen(Sande) « Medlem Jon Henrik Grindlia (Andebu) « Medlem Terje Fuglevik(Nøtterøy) « Medlem Gunnar Akerholt (Hof) « Medlem Anne Marie Indseth (Tjøme) « Medlem Hanna Therese Berg (Holmestr) « Medlem Ingar Døler (Larvik) « Medlem Kjetil Holm Klavenes( vfk) « Medlem Anders Assev (Lardal) « Medlem Frode Hestnes (Re) « Vara Are Karlsen (Horten) « Vara Marianne Teisbekk (Tønsberg) « Vara Paul Gregersen (Sande « Vara Frank Vikansen (Andebu) « Vara Aud Eiken (Nøtterøy) « Vara Arve Stordal (Hof) « Vara Dag Erichsrud (Tjøme) « Vara Hans Petter Harestad (Holmest)) « Vara Live G. Jetlund (Larvik) « Vara Rune Mathiassen (k) « Va ra Kari Stensholt (Lardal) « Vara Harald Solberg (Re) « Daglig leder, lønn og godtgjørelse Navn Årslønn kr Godtgjørelse i 2014 Orrvar Dalby 695.250 0 Økonomiske nøkkeltall Drift 2014 Kr -
Norway Maps.Pdf
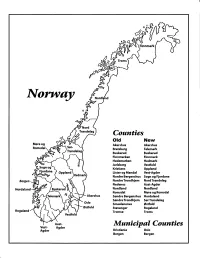
Finnmark lVorwny Trondelag Counties old New Akershus Akershus Bratsberg Telemark Buskerud Buskerud Finnmarken Finnmark Hedemarken Hedmark Jarlsberg Vestfold Kristians Oppland Oppland Lister og Mandal Vest-Agder Nordre Bergenshus Sogn og Fjordane NordreTrondhjem NordTrondelag Nedenes Aust-Agder Nordland Nordland Romsdal Mgre og Romsdal Akershus Sgndre Bergenshus Hordaland SsndreTrondhjem SorTrondelag Oslo Smaalenenes Ostfold Ostfold Stavanger Rogaland Rogaland Tromso Troms Vestfold Aust- Municipal Counties Vest- Agder Agder Kristiania Oslo Bergen Bergen A Feiring ((r Hurdal /\Langset /, \ Alc,ersltus Eidsvoll og Oslo Bjorke \ \\ r- -// Nannestad Heni ,Gi'erdrum Lilliestrom {", {udenes\ ,/\ Aurpkog )Y' ,\ I :' 'lv- '/t:ri \r*r/ t *) I ,I odfltisard l,t Enebakk Nordbv { Frog ) L-[--h il 6- As xrarctaa bak I { ':-\ I Vestby Hvitsten 'ca{a", 'l 4 ,- Holen :\saner Aust-Agder Valle 6rrl-1\ r--- Hylestad l- Austad 7/ Sandes - ,t'r ,'-' aa Gjovdal -.\. '\.-- ! Tovdal ,V-u-/ Vegarshei I *r""i'9^ _t Amli Risor -Ytre ,/ Ssndel Holt vtdestran \ -'ar^/Froland lveland ffi Bergen E- o;l'.t r 'aa*rrra- I t T ]***,,.\ I BYFJORDEN srl ffitt\ --- I 9r Mulen €'r A I t \ t Krohnengen Nordnest Fjellet \ XfC KORSKIRKEN t Nostet "r. I igvono i Leitet I Dokken DOMKIRKEN Dar;sird\ W \ - cyu8npris Lappen LAKSEVAG 'I Uran ,t' \ r-r -,4egry,*T-* \ ilJ]' *.,, Legdene ,rrf\t llruoAs \ o Kirstianborg ,'t? FYLLINGSDALEN {lil};h;h';ltft t)\l/ I t ,a o ff ui Mannasverkl , I t I t /_l-, Fjosanger I ,r-tJ 1r,7" N.fl.nd I r\a ,, , i, I, ,- Buslr,rrud I I N-(f i t\torbo \) l,/ Nes l-t' I J Viker -- l^ -- ---{a - tc')rt"- i Vtre Adal -o-r Uvdal ) Hgnefoss Y':TTS Tryistr-and Sigdal Veggli oJ Rollag ,y Lvnqdal J .--l/Tranbv *\, Frogn6r.tr Flesberg ; \. -

Jordskifterett
NEDRE TELEMARK JORDSKIFTERETT Fylkesmannen i Telemark v/Johan Aas Postboks 2603 3702 SKIEN Saksnummer Vår referanse Vår dato 0800-201 I-0005 1136/12gw 01.06.2012 Forkynning av rettsbok Sak 0800-2011-0005 VINDFJELL naturreservat ved Nedre Telemark jordskifterett ble avsluttet 01.06.2012. Rettsboka i saken er vedlagt og regnes som forkynt 08.06.2012. At dokumentet er forkynt, vil si at det er kommet fram til deg, slik at du kan gjøre deg kjent med innholdet. Forkynningsdatoen ovenfor er utgangspunkt for beregning av ankefrister m.m. Dersom du mottar dette brevet seinere enn datoen som er nevnt over og ønsker å gjøre dette gjeldende, må du straks melde fra til vårt kontor. Årsaken til forsinkelsen må da opplyses. Ankefristen er en måned regnet fra forkynningsdatoen. Spørsmål angående forkynningen kan tas opp over telefon eller ved personlig henvendelse til vårt kontor. Vennligst bekreft at brevet er mottatt ved å undertegne og returnere svarslippen i vedlagte frankerte konvolutt snarest, helst innen fem dager. Dersom du skylder sakskostnader, ber vi om at de betales innen fristen. Utfylt innbetalings- blankett er vedlagt. Med hilsen Gunnar Roe overingeniør Ty:direkte innvalg: 21 55 32 96 E-post: gunnarroe@domsto1no Vedlegg mottakskvittering - rettsbok kopi av jordskiftekart - kopi av jordskifteloven kap. 7 - rettsmiddel - innbetalingsblankett til parter som skylder sakskostnader adresseliste Postadresse Besoksadresse Telefon 21 55 33 00 Bank 7874 06 85049 Postboks 2845 Fylkesbakken 10 Telefax Bedriftsnr. 974 730 995 3702 Skien 3715 Skien jskipos6aldomstolno www.dornstol.no/jski Nedre Telemark jordskifterett Rettsbok Sak: 0800-2011-0005 VINDFJELL naturreservat Gnr. 71, 72, 73, 74 og 75 i Lardal kommune og Gnr. -

Fastlegetest Vestfold
Rang Rang Antall Snitt pr Generell Konkret Konkret Digitale landet fylket FASTLEGEKONTOR Kommune leger liste info info service tjenester SUM 3 1 Hof Legekontor Hof 2 1 200 100,0 50,0 100,0 80,0 74 25 2 Kirkebygden Legekontor Tjøme 1 1 200 76,0 51,0 100,0 65,0 67,5 36 3 Gleditschgården Legesenter As Sandefjord 4 1 140 100,0 50,0 100,0 55,0 66,5 40 4 Legegruppen As Larvik 4 1 150 96,0 60,0 94,0 45,0 65,9 41 5 Brygga Legekontor Tønsberg Tønsberg 3 1 567 96,0 49,0 100,0 55,0 65,7 63 6 Revetal Legekontor Re 2 1 325 80,0 60,0 94,0 45,0 64,3 64 7 Bellevue Legesenter Nøtterøy 2 1 400 100,0 55,0 94,0 45,0 64,3 125 8 Vestskogen Medisinske Senter Nøtterøy 3 1500 80 54 94 45 61,9 140 9 Nordbyen Legesenter Haugvik As Larvik 3 1450 100 51 88 45 61,5 141 10 Våle Legekontor Re 4 1125 76 60 82 45 61,5 201 11 Legekontoret På Tolvsrød Tønsberg 3 1300 76 51 94 45 60,3 219 12 Re Legegruppe Da Re 4 713 76 50 94 45 59,9 256 13 3 Leger Tønsberg 3 1400 76 48 94 45 59,1 273 14 Helgeroa Legesenter Da Larvik 2 1425 76 50 88 45 58,7 288 15 Familielegen As Sandefjord 2 1300 96 49 60 57,5 58,5 296 16 Legekontoret På Tjøme Tjøme 3 995 96 50 76 45 58,3 311 17 Kilen Legesenter Da Sandefjord 3 1150 96 63,5 48 45 58,1 361 18 Midtløkken Legekontor Tønsberg 3 1258 96 44,5 82 45 57,3 368 19 Lardal Legekontor Lardal 2 1305 36 58 54 65 57,1 413 20 Åsgårdstrand Helsesenter Horten 5 1560 100 50 48 55 56,1 414 21 Sem Legekontor Tønsberg 1 2000 76 56 48 55 56,1 433 22 Åsgårdstrand Legekontor Horten 3 1387 76 38,5 82 55 55,9 444 23 Byskogen Legesenter Larvik 1 2000 80 60 54 42,5 55,6 -

Hedrum Og Lardal
HEDRUM OG LARDAL NR. 3 - 2018 • Å R G A N G 7 SE OGSÅ VÅRE HJEMMESIDER: www.larvik.kirken.no 2 • KONTAKTINFORMASJON INNHOLD Hedrum menighet: Leder i menighetsrådet: Kjell Østby mobil 982 31 266 Til våre lesere 4 Kirketjener/klokker: Astrid Helene Floen mobil 938 44 037 Andakt 5 Kateket: Lasse Rasmussen Moskvil mobil 467 98 879 Min salme 6 Kvelde menighet: Bekjemp vold mot kvinner i Nepal 7 Leder i menighetsrådet: Vibeke Kvalevåg mobil 456 04 526 Barnedåp i Hvarnes-skauen 8 Kirketjener/klokker: Morten Meidell-Pritzier Hagelund mobil 913 81 815 Flott Olsokkfeiring i Hedrum 9 Barne- og ungdomsarbeider: Karoline Bredvei Hvarnes mobil 473 99 414 Spellemannen 10 Hvarnes menighet: Konfirmantene i Lardal 12 Leder i menighetsrådet: Geir Bårnes mobil 952 81 991 Diamantkonfirmantene i Lardal 13 Kirketjener/klokker: Marit Thorvaldsen mobil 920 99 703 Saniteten og kvinnehelse 14 Barne- og ungdomsarbeider: Karoline Bredvei Hvarnes mobil 473 99 414 Fra orgelkrakken 15 Kvelde Åpen Barnehage 16 Lardal menighet: Sponsorsider 18 Leder i menighetsrådet: Inger Lene Røsholt mobil 977 03 971 Besøk på lysstøperi 20 Kirketjener Hem kirke, og leder for kirkegårdsdelen i Lardal: Kvelde og Hvarnes menigheter inviterer til familiemiddag 21 Jørn Olav Lie mobil 916 27 537 Dette skjer i Kvelde 22 Kirketjener i Svarstad kirke: Kai Berntsen mobil 932 49 183 Dette skjer i Hvarnes 23 Kirketjener i Styrvoll kirke: Annette Gran mobil 910 07 987 Dette skjer i Hedrum 24 Menighetspedagog/diakoniarbeider: Mariann Eskedal mobil 971 99 722 Dette skjer i Lardal 25 IKT-ansvarlig: -

Kommune- Og Regionreformene
Kommune- og regionreformene Teknisk koordinator på IKT- spørsmål Over 50 år siden sist vi hadde kommunereform 1964 2017 Agenda 1. Bakgrunn for Kartverkets rolle 2. Vedtatte sammenslåinger 3. Erfaringer og utfordringsbildet 4. Hvordan jobber Kartverket 5. Hvordan skal kommunene forberede seg Bakgrunn for Kartverkets rolle – erfaringer etter innføring av matrikkelen • Inderøy og Mosvik 2012 • Harstad og Bjarkøy 2013 • Sandefjord, Andebu og Stokke (SAS) 2017 • Larvik – Lardal, Holmestrand – Hof, Tjøme – Nøtterøy og Rissa – Leksvik • Nord- og Sør- Trøndelag • Alt har gått bra matrikkelmessig. • Sammenslåing griper inn i mange IKT systemer. • Behov for nasjonal samordning. Matrikkelen, ett av tre nasjonale basisregistre • Folkeregisteret (grunndata om person – hvem?) • Enhetsregisteret (grunndata om virksomhet – hva?) • Matrikkelen (grunndata om eiendom – hvor?) Illustrasjonen viser dataflyten inn og ut av matrikkelen, og er ikke uttømmende. Tildelingsbrev fra KMD • Oppdrag 3.3.7: • Oppdrag 3.3.8: Innhold i Kartverkets rolle 1) Som teknisk koordinator skal Kartverket: • Synliggjøre og aktualisere utfordringene som ligger i IKT-systemer for offentlig sektor i forbindelse med reformene. • Koordinere at store statlige aktører tilpasser utvikler sine IKT-systemer til å takle reformen. • Skal synliggjøre HVA som må gjøres og NÅR - Ikke HVORDAN. • Gjelder alle fagområder i kommunen – IKT-utfordringer. • Kartverket skal ha statusoversikt på overordnet nivå. • Belyse utfordringsbildet. • Vise vei til verktøykassen – kartverket.no. • Koordinere og kanalisere spørsmål - fagetater følger opp sitt fagområde. 2) Som faglig støtte skal Kartverket: • Gi faglig støtte innenfor fagområdet matrikkel. Overordnet • Kommune- og fylkessammenslåinger gjennomføres ALLTID 1. januar • Kommunenummer gjeldende fra 2020 er klare, og kommuner har fått brev fra KMD • Ny kommune = ny juridisk organisasjon. Gjennomgang/ reforhandling av alle juridiske avtaler opp mot leverandører Vedtak og veien videre Stortingsvedtak 8. -

Naturvernområder I Vestfold Naturreservater Omfatter Områder Med Uberørt Eller Lite Berørt Natur, Eller Utgjør En Spesiell Naturtype
Adalstjern, HORTEN Sønstegård, TJØME Rød-Dirhue, TJØME Gullkronene, TØNSBERG Kommersøya, SANDE Grunnane, SVELVIK Strandvika, SANDEFJORD Naturvernområder i Vestfold Naturreservater omfatter områder med uberørt eller lite berørt natur, eller utgjør en spesiell naturtype. Verneformen er den strengeste etter Oslofjordens geologi, klima og landskap gir naturvernloven og naturmangfoldloven. grunnlag for et rikt mangfold av planter Naturminner er knyttet til trefredninger, mineraler, fossiler eller og dyr. 155 områder og objekter kvartærgeologiske forekomster. (geologiske forekomster og trær) i Biotopvern brukes for å beskytte leveområder for planter og dyr. Vestfold er vernet. Verneområdene Verneformen anvendes ofte på mindre områder eller områder som av utgjør i overkant av to prosent av ulike grunner ikke passer som naturreservat. fylkets landareal. Formålet med Landskapsvernområder omfatter egenartede eller vakre natur- eller kulturlandskap. Verneformen brukes ofte for å ta vare på å verne områder er å ta vare kulturlandskap i aktiv bruk. Verneformen er mildere enn de øvrige på et utvalg sjeldne, sårbare, verneformene. urørte og typiske landskap og Områder i Ormø-Færder landskapsvernområde der det er et mål å bevare det egenartede kulturlandskapet, preget av øygårdsbruk og naturmiljøer samt plante- og tidligere bosetting. dyrearter. Områder i Ormø-Færder landskapsvernområde der det er et mål å bevare et spesielt plante- og dyreliv. Områder i Ormø-Færder landskapsvernområde der det er et mål å beskytte sjøfuglene mot forstyrrelser i hekketiden. N V Ø S 1:230 000 0 10 km Tillatelsesnummer Statens kartverk: Mad12002 - R127454 Tillatelsesnummer GEOVEKST: GV-L-0700 Lisensnr.: 12247NE-540741 OPPSYN: Statens Naturoppsyn Vestfold, www.dirnat.no Utgitt av Fylkesmannen i Vestfold mars 2010 FORVALTNING OG SKJØTSEL: Fylkesmannen i Vestfold, www.fylkesmannen.no Formgivning Bibro Design Naturvernområder i Vestfold - 2 SVELVIK 29.