Supplementary Table S1. Summary of MEROPS and ESTHER Database Description for Peptidases and Lipases, Respectively
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Screening and Identification of Key Biomarkers in Clear Cell Renal Cell Carcinoma Based on Bioinformatics Analysis
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Screening and identification of key biomarkers in clear cell renal cell carcinoma based on bioinformatics analysis Basavaraj Vastrad1, Chanabasayya Vastrad*2 , Iranna Kotturshetti 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. 3. Department of Ayurveda, Rajiv Gandhi Education Society`s Ayurvedic Medical College, Ron, Karnataka 562209, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Clear cell renal cell carcinoma (ccRCC) is one of the most common types of malignancy of the urinary system. The pathogenesis and effective diagnosis of ccRCC have become popular topics for research in the previous decade. In the current study, an integrated bioinformatics analysis was performed to identify core genes associated in ccRCC. An expression dataset (GSE105261) was downloaded from the Gene Expression Omnibus database, and included 26 ccRCC and 9 normal kideny samples. Assessment of the microarray dataset led to the recognition of differentially expressed genes (DEGs), which was subsequently used for pathway and gene ontology (GO) enrichment analysis. -

A Review on Agmatinase Inhibitors
www.ijcrt.org © 2020 IJCRT | Volume 8, Issue 12 December 2020 | ISSN: 2320-2882 A review on Agmatinase inhibitors 1Sunnica Biswas, 2Mr. R. T. Lohiya, 3Dr. Milind Umekar *1&2 Department Of Pharmaceutical Chemistry Smt. Kishoriti Bhoyar College of Pharmacy, Kamptee, Dst. Nagpur, 441002 Abstract : Agmatine is the product of arginine decarboxylation and can be hydrolyzed by agmatinase to putrescine, the precursor for biosynthesis of higher polyamines, spermidine, and spermine. Besides being an intermediate in polyamine metabolism, recent findings indicate that agmatine may play important regulatory roles in mammals. Agmatine, 4-aminobutyl guanidine, has recently been found in various mammalian organs and is thought to act as a neurotransmitter or neuromodulatory agent. The present study is to do a review on agmatine and its synthesized analogues till now for agmatinase inhibitory action. Agmatinase is a binuclear manganese metalloenzyme and belongs to the ureohydrolase superfamily that includes arginase, formiminoglutamase, and proclavaminate amidinohydrolase. Compared with a wealth of structural information available for arginases, no three dimensional structure of agmatinase has been reported. Agmatinase is an enzyme which blocks the mammalian agmatine which is ultimately responsible for the agmatine degradation in the body. Agmatinase is an enzyme which regulates the half life of agmatine in the brain. Hence a selective inhibitor of brain agmatinase is required. Several derivatives of agmatine are synthesized previously for agmatinase inhibitory activity but none of them showed selective inhibition. PZC (Piperazinecarboxamidine) is a derivative of agmatine or guanidine is expected to show selective inhibition of human agmatinase. A detailed review is carried out in order to understand the agmatinase inhibitor. -

Quantum Effects in Radical B12 Enzymes
Quantum Effects in Adenosylcobalamin-dependent Enzymes by M. Hossein Khalilian Boroujeni B.Sc., Chemistry, Razi University, 2014 A THESIS SUBMITTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF MASTER OF SCIENCE in THE COLLEGE OF GRADUATE STUDIES (Chemistry) THE UNIVERSITY OF BRITISH COLUMBIA (Okanagan) April 2019 © M. Hossein Khalilian Boroujeni, 2019 The following individuals certify that they have read, and recommend to the College of Graduate Studies for acceptance, a thesis/dissertation entitled: Quantum Effects in Adenosylcobalamin-dependent Enzymes submitted by M. Hossein Khalilian Boroujeni in partial fulfillment of the requirements for the degree of Master of Science Examining Committee: Gino A. DiLabio, I. K. Barber School of Arts & Sciences Supervisor W. Stephen McNeil, I. K. Barber School of Arts & Sciences Supervisory Committee Member Kirsten Wolthers, I. K. Barber School of Arts & Sciences Supervisory Committee Member Michael Deyholos, I. K. Barber School of Arts & Sciences University Examiner ii Abstract The ability of radical enzymes to maintain tight control over the high reactive radical intermediates generated in their active sites is not completely understood. In this thesis, we report on a strategy that radical (B12-dependent) enzymes appear to exploit in order to manipulate and control the reactivity of one of their radical intermediate (5'-deoxyadenosyl radical) contained in the active site. The results of quantum mechanical calculations suggest that these enzymes utilize the little known quantum Coulombic effect (QCE), which causes the radical to acquire an electronic structure that contradicts the Aufbau Principle. This effect causes the energy of the singly-occupied molecular orbital (SOMO) of the radical to be well below that of the highest-occupied molecular orbital (HOMO), which renders the radical less reactive. -

Agmatinase Sirna (H): Sc-60060
SANTA CRUZ BIOTECHNOLOGY, INC. Agmatinase siRNA (h): sc-60060 BACKGROUND STORAGE AND RESUSPENSION Agmatinase (also known as agmatine ureohydrolase) results from the decar- Store lyophilized siRNA duplex at -20° C with desiccant. Stable for at least boxylation of L-arginine by arginine decarboxylase to form a metabolic inter- one year from the date of shipment. Once resuspended, store at -20° C, mediate in the biosynthesis of putresine and higher polyamines (spermidine avoid contact with RNAses and repeated freeze thaw cycles. and spermine). Agmatinase has been shown to play a role in several important Resuspend lyophilized siRNA duplex in 330 µl of the RNAse-free water biochemical processes in humans, ranging from effects on the central nervous provided. Resuspension of the siRNA duplex in 330 µl of RNAse-free water system to cell proliferation in cancer and viral replication. Agmatinase cat- makes a 10 µM solution in a 10 µM Tris-HCl, pH 8.0, 20 mM NaCl, 1 mM alyzes the hydrolysis of agmatine to putresine and urea and is a major target EDTA buffered solution. for drug therapy. Human Agmatinase retains about 30% identity to bacterial agmatinases and less than 20% identity to mammalian arginases. Residues APPLICATIONS required for binding of Mn2+ at the active site in bacterial Agmatinase and other members of the arginase superfamily are fully conserved in human Agmatinase siRNA (h) is recommended for the inhibition of Agmatinase Agmatinase. Agmatinase mRNA is most abundant in human liver and kidney, expression in human cells. but is also expressed in several other tissues, including skeletal muscle and brain. -

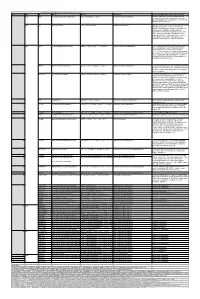
Table 2. Significant
Table 2. Significant (Q < 0.05 and |d | > 0.5) transcripts from the meta-analysis Gene Chr Mb Gene Name Affy ProbeSet cDNA_IDs d HAP/LAP d HAP/LAP d d IS Average d Ztest P values Q-value Symbol ID (study #5) 1 2 STS B2m 2 122 beta-2 microglobulin 1452428_a_at AI848245 1.75334941 4 3.2 4 3.2316485 1.07398E-09 5.69E-08 Man2b1 8 84.4 mannosidase 2, alpha B1 1416340_a_at H4049B01 3.75722111 3.87309653 2.1 1.6 2.84852656 5.32443E-07 1.58E-05 1110032A03Rik 9 50.9 RIKEN cDNA 1110032A03 gene 1417211_a_at H4035E05 4 1.66015788 4 1.7 2.82772795 2.94266E-05 0.000527 NA 9 48.5 --- 1456111_at 3.43701477 1.85785922 4 2 2.8237185 9.97969E-08 3.48E-06 Scn4b 9 45.3 Sodium channel, type IV, beta 1434008_at AI844796 3.79536664 1.63774235 3.3 2.3 2.75319499 1.48057E-08 6.21E-07 polypeptide Gadd45gip1 8 84.1 RIKEN cDNA 2310040G17 gene 1417619_at 4 3.38875643 1.4 2 2.69163229 8.84279E-06 0.0001904 BC056474 15 12.1 Mus musculus cDNA clone 1424117_at H3030A06 3.95752801 2.42838452 1.9 2.2 2.62132809 1.3344E-08 5.66E-07 MGC:67360 IMAGE:6823629, complete cds NA 4 153 guanine nucleotide binding protein, 1454696_at -3.46081884 -4 -1.3 -1.6 -2.6026947 8.58458E-05 0.0012617 beta 1 Gnb1 4 153 guanine nucleotide binding protein, 1417432_a_at H3094D02 -3.13334396 -4 -1.6 -1.7 -2.5946297 1.04542E-05 0.0002202 beta 1 Gadd45gip1 8 84.1 RAD23a homolog (S. -

Development of a Phage Display Library for Discovery of Antigenic Brucella Peptides Jeffrey Williams Iowa State University
Iowa State University Capstones, Theses and Graduate Theses and Dissertations Dissertations 2018 Development of a phage display library for discovery of antigenic Brucella peptides Jeffrey Williams Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/etd Part of the Microbiology Commons Recommended Citation Williams, Jeffrey, "Development of a phage display library for discovery of antigenic Brucella peptides" (2018). Graduate Theses and Dissertations. 16896. https://lib.dr.iastate.edu/etd/16896 This Thesis is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. Development of a phage display library for discovery of antigenic Brucella peptides by Jeffrey Williams A thesis submitted to the graduate faculty in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE Major: Microbiology Program of Study Committee: Bryan H. Bellaire, Major Professor Steven Olsen Steven Carlson The student author, whose presentation of the scholarship herein was approved by the program of study committee, is solely responsible for the content of this thesis. The Graduate College will ensure this thesis is globally accessible and will not permit alterations after a degree is conferred. Iowa State University -

Anaerobic Radical Enzymes for Biotechnology
ChemBioEng Reviews Anaerobic radical enzymes for biotechnology Journal: ChemBioEng Reviews Manuscript ID cben.201800003.R1 Wiley - Manuscript type:For Review Peer Review Date Submitted by the Author: n/a Complete List of Authors: Jäger, Christof; University of Nottingham, Chemical and Environmental Engineering Croft, Anna; University of Nottingham, Chemical and Environmental Engineering Keywords: Radicals, Enzymes, Catalysis, Biotechnology, Anaerobic reactions Wiley-VCH Page 1 of 61 ChemBioEng Reviews 1 2 3 Christof M. Jäger* and Anna K. Croft* 4 5 6 7 8 9 Anaerobic radical enzymes for biotechnology 10 11 12 13 AUTHORS: Dr Christof Martin Jäger* and Dr Anna Kristina Croft* 14 15 16 ADDRESS: Department of Chemical and Environmental Engineering, University of 17 18 Nottingham, Nottingham, NG7 2RD, United Kingdom. [email protected], 19 For Peer Review 20 21 [email protected] 22 23 24 ABSTRACT: 25 26 27 28 Enzymes that proceed through radical intermediates have a rich chemistry that includes 29 30 functionalisation of otherwise unreactive carbon atoms, carbon-skeleton rearrangements, 31 32 aromatic reductions, and unusual eliminations. Especially under anaerobic conditions, 33 34 organisms have developed a wide range of approaches for managing these transformations 35 36 that can be exploited to generate new biological routes towards both bulk and specialty 37 38 39 chemicals. These routes are often either much more direct or allow access to molecules that 40 41 are inaccessible through standard (bio)chemical approaches. This review gives an overview 42 43 of some of the key enzymes in this area: benzoyl-CoA reductases (that effect the enzymatic 44 45 Birch reduction), ketyl radical dehydratases, coenzyme B12-dependant enzymes, glycyl 46 47 radical enzymes, and radical SAM (AdoMet radical) enzymes. -
When the Reaction Is
Table S3. iJL1678-ME model modification (blocked reactions) Iter. Cat. ID Name Formula Subsystem Comments (When the reaction is turned on) 1 bp2 EDD 6-phosphogluconate dehydratase 6pgc_c⇌2ddg6p_c + h2o_c Pentose Phosphate Pathway Create a major effect of steep acetate overflow elevation in high growth. Comparing to the main glycolytic pathway, it is metabolicly less efficient but proteomicly more efficient. bp1 ICL Isocitrate lyase icit_c→glx_c + succ_c Anaplerotic Reactions Bypass for the main TCA cycle pathways from turning isocitrate to succinate, when ICL is turned on, Isocitrate dehydrogenase(ICDHyr), 2-Oxogluterate dehydrogenase(AKGDH) and Succinyl-CoA synthetase (ATP-forming,SUCOAS) would reduce. Ref. (1) and (2) shows that this reaction is off in higher growth. Ref. (3) shows that this reaction is converging to being off when the dynamic of respiration using enzyme kinetics is simulated. 2 bp1 ABTA 4-aminobutyrate transaminase 4abut_c + akg_c⇌glu__L_c + sucsal_c Arginine and Proline Metabolism Another backup pathway of succinate production, from 2-Oxoglutarate (akg). Respiration would be induced when it is on, since the flux through ETC(CYTBO3_4pp and ATPS4rpp) would increase. As it requires the co-factor pyridoxal 5'-phosphate(2−) to get catalyzed(4), indicating that this reaction is regulated by the flux of other reactions(pyridoxal 5'- phosphate(2-) production, etc.). 3 GLYAT Glycine C-acetyltransferase accoa_c + gly_c⇌2aobut_c + coa_c Glycine and Serine Metabolism A reaction that back up for the respiration. Reactions fluxes in TCA cycle would drop when this reaction is turned on. It also requires pyridoxal 5'-phosphate(2−) for the regulation. 4 NADTRHD NAD transhydrogenase nad_c + nadph_c⇌nadh_c + nadp_c Oxidative Phosphorylation A reaction that would make the transition between NAD and NADP metabolically more efficient. -

Characterization of the Scavenger Cell Proteome in Mouse and Rat Liver
Biol. Chem. 2021; 402(9): 1073–1085 Martha Paluschinski, Cheng Jun Jin, Natalia Qvartskhava, Boris Görg, Marianne Wammers, Judith Lang, Karl Lang, Gereon Poschmann, Kai Stühler and Dieter Häussinger* Characterization of the scavenger cell proteome in mouse and rat liver + https://doi.org/10.1515/hsz-2021-0123 The data suggest that the population of perivenous GS Received January 25, 2021; accepted July 4, 2021; scavenger cells is heterogeneous and not uniform as previ- published online July 30, 2021 ously suggested which may reflect a functional heterogeneity, possibly relevant for liver regeneration. Abstract: The structural-functional organization of ammonia and glutamine metabolism in the liver acinus involves highly Keywords: glutaminase; glutamine synthetase; liver specialized hepatocyte subpopulations like glutamine syn- zonation; proteomics; scavenger cells. thetase (GS) expressing perivenous hepatocytes (scavenger cells). However, this cell population has not yet been char- acterized extensively regarding expression of other genes and Introduction potential subpopulations. This was investigated in the present study by proteome profiling of periportal GS-negative and There is a sophisticated structural-functional organization in perivenous GS-expressing hepatocytes from mouse and rat. the liver acinus with regard to ammonium and glutamine Apart from established markers of GS+ hepatocytes such as metabolism (Frieg et al. 2021; Gebhardt and Mecke 1983; glutamate/aspartate transporter II (GLT1) or ammonium Häussinger 1983, 1990). Periportal hepatocytes express en- transporter Rh type B (RhBG), we identified novel scavenger zymes required for urea synthesis such as the rate-controlling cell-specific proteins like basal transcription factor 3 (BTF3) enzyme carbamoylphosphate synthetase 1 (CPS1) and liver- and heat-shock protein 25 (HSP25). -

Generate Metabolic Map Poster
Authors: Pallavi Subhraveti Ron Caspi Peter Midford Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_001591825Cyc: Bacillus vietnamensis NBRC 101237 Cellular Overview Connections between pathways are omitted for legibility. Anamika Kothari sn-glycerol phosphate phosphate pro phosphate phosphate phosphate thiamine molybdate D-xylose D-ribose glutathione 3-phosphate D-mannitol L-cystine L-djenkolate lanthionine α,β-trehalose phosphate phosphate [+ 3 more] α,α-trehalose predicted predicted ABC ABC FliY ThiT XylF RbsB RS10935 UgpC TreP PutP RS10200 PstB PstB RS10385 RS03335 RS20030 RS19075 transporter transporter of molybdate of phosphate α,β-trehalose 6-phosphate L-cystine D-xylose D-ribose sn-glycerol D-mannitol phosphate phosphate thiamine glutathione α α phosphate phosphate phosphate phosphate L-djenkolate 3-phosphate , -trehalose 6-phosphate pro 1-phosphate lanthionine molybdate phosphate [+ 3 more] Metabolic Regulator Amino Acid Degradation Amine and Polyamine Biosynthesis Macromolecule Modification tRNA-uridine 2-thiolation Degradation ATP biosynthesis a mature peptidoglycan a nascent β an N-terminal- -

(10) Patent No.: US 8119385 B2
US008119385B2 (12) United States Patent (10) Patent No.: US 8,119,385 B2 Mathur et al. (45) Date of Patent: Feb. 21, 2012 (54) NUCLEICACIDS AND PROTEINS AND (52) U.S. Cl. ........................................ 435/212:530/350 METHODS FOR MAKING AND USING THEMI (58) Field of Classification Search ........................ None (75) Inventors: Eric J. Mathur, San Diego, CA (US); See application file for complete search history. Cathy Chang, San Diego, CA (US) (56) References Cited (73) Assignee: BP Corporation North America Inc., Houston, TX (US) OTHER PUBLICATIONS c Mount, Bioinformatics, Cold Spring Harbor Press, Cold Spring Har (*) Notice: Subject to any disclaimer, the term of this bor New York, 2001, pp. 382-393.* patent is extended or adjusted under 35 Spencer et al., “Whole-Genome Sequence Variation among Multiple U.S.C. 154(b) by 689 days. Isolates of Pseudomonas aeruginosa” J. Bacteriol. (2003) 185: 1316 1325. (21) Appl. No.: 11/817,403 Database Sequence GenBank Accession No. BZ569932 Dec. 17. 1-1. 2002. (22) PCT Fled: Mar. 3, 2006 Omiecinski et al., “Epoxide Hydrolase-Polymorphism and role in (86). PCT No.: PCT/US2OO6/OOT642 toxicology” Toxicol. Lett. (2000) 1.12: 365-370. S371 (c)(1), * cited by examiner (2), (4) Date: May 7, 2008 Primary Examiner — James Martinell (87) PCT Pub. No.: WO2006/096527 (74) Attorney, Agent, or Firm — Kalim S. Fuzail PCT Pub. Date: Sep. 14, 2006 (57) ABSTRACT (65) Prior Publication Data The invention provides polypeptides, including enzymes, structural proteins and binding proteins, polynucleotides US 201O/OO11456A1 Jan. 14, 2010 encoding these polypeptides, and methods of making and using these polynucleotides and polypeptides. -

Microbial Biochemistry, 2Nd Edition
Microbial Biochemistry . G.N. Cohen Microbial Biochemistry Second Edition Prof. G.N. Cohen Institut Pasteur rue du Docteur Roux 28 75724 Paris France [email protected] ISBN 978-90-481-9436-0 e-ISBN 978-90-481-9437-7 DOI 10.1007/978-90-481-9437-7 Springer Dordrecht Heidelberg London New York Library of Congress Control Number: 2010938472 # Springer Science+Business Media B.V. 2011 No part of this work may be reproduced, stored in a retrieval system, or transmitted in any form or by any means, electronic, mechanical, photocopying, microfilming, recording or otherwise, without written permission from the Publisher, with the exception of any material supplied specifically for the purpose of being entered and executed on a computer system, for exclusive use by the purchaser of the work. Printed on acid-free paper Springer is part of Springer Science+Business Media (www.springer.com) Foreword This book originates from almost 60 years of living in the company of micro- organisms, mainly with Escherichia coli. My scientific life has taken place almost exclusively at the Institut Pasteur in Paris, where many concepts of modern molecular biology were born or developed. The present work emphasizes the interest of microbial physiology, biochemistry and genetics. It takes into account the considerable advances which have been made in the field in the last 30 years by the introduction of gene cloning and sequencing and by the exponential development of physical methods such as X-ray crystallog- raphy of proteins. The younger generation of biochemists is legitimately interested in the problems raised by differentiation and development in higher organisms, and also in neuros- ciences.