Genomic Evidence of Reactive Oxygen Species Elevation in Papillary Thyroid Carcinoma with Hashimoto Thyroiditis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

OXSR1 (A-4): Sc-271707
SANTA CRUZ BIOTECHNOLOGY, INC. OXSR1 (A-4): sc-271707 BACKGROUND APPLICATIONS Oxidative stress-responsive 1 protein (OXSR1), a protein of 527 amino acids, OXSR1 (A-4) is recommended for detection of OXSR1 of mouse, rat and belongs to the STE20 subfamily. OXSR1 is one of two human homologs of human origin by Western Blotting (starting dilution 1:100, dilution range Fray, a serine/threonine kinase expressed in Drosophila. OXSR1 binds to and 1:100-1:1000), immunoprecipitation [1-2 µg per 100-500 µg of total protein phosphorylates p21-activated protein kinase (PAK1) and regulates downstream (1 ml of cell lysate)], immunofluorescence (starting dilution 1:50, dilution kinases in response to environmental stress. Endogenous OXSR1 is activated range 1:50-1:500) and solid phase ELISA (starting dilution 1:30, dilution only by osmotic stresses, notably sorbitol and to a lesser extent NaCl. OXSR1 range 1:30-1:3000). may also play a role in regulating the Actin cytoskeleton. The chloride channel Suitable for use as control antibody for OXSR1 siRNA (h): sc-61273, OXSR1 proteins SLC12A1, SLC12A2 and SLC12A6 isoform 2 interact with OXSR1, siRNA (m): sc-61274, OXSR1 shRNA Plasmid (h): sc-61273-SH, OXSR1 but SLC12A4 and SLC12A7 do not. The WNK1 and WNK4 protein kinases shRNA Plasmid (m): sc-61274-SH, OXSR1 shRNA (h) Lentiviral Particles: activate OXSR1 by phosphorlating its T-loop. The OXSR1 protein is widely sc-61273-V and OXSR1 shRNA (m) Lentiviral Particles: sc-61274-V. expressed in mammalian tissues. Molecular Weight of OXSR1: 58 kDa. REFERENCES Positive Controls: OXSR1 (h3): 293 Lysate: sc-158793, HeLa whole cell lysate: 1. -

Novel Binding Partners of PBF in Thyroid Tumourigenesis
NOVEL BINDING PARTNERS OF PBF IN THYROID TUMOURIGENESIS By Neil Sharma A thesis presented to the College of Medical and Dental Sciences at the University of Birmingham for the Degree of Doctor of Philosophy Centre for Endocrinology, Diabetes and Metabolism, School of Clinical and Experimental Medicine August 2013 University of Birmingham Research Archive e-theses repository This unpublished thesis/dissertation is copyright of the author and/or third parties. The intellectual property rights of the author or third parties in respect of this work are as defined by The Copyright Designs and Patents Act 1988 or as modified by any successor legislation. Any use made of information contained in this thesis/dissertation must be in accordance with that legislation and must be properly acknowledged. Further distribution or reproduction in any format is prohibited without the permission of the copyright holder. SUMMARY Thyroid cancer is the most common endocrine cancer, with a rising incidence. The proto-oncogene PBF is over-expressed in thyroid tumours, and the degree of over-expression is directly linked to patient survival. PBF causes transformation in vitro and tumourigenesis in vivo, with PBF-transgenic mice developing large, macro-follicular goitres, effects partly mediated by the internalisation and repression of the membrane-bound transporters NIS and MCT8. NIS repression leads to a reduction in iodide uptake, which may negatively affect the efficacy of radioiodine treatment, and therefore prognosis. Work within this thesis describes the use of tandem mass spectrometry to produce a list of potential binding partners of PBF. This will aid further research into the pathophysiology of PBF, not just in relation to thyroid cancer but also other malignancies. -

(MMP-9) in Oral Cancer Squamous Cells—Are There Therapeutical Hopes?
materials Article Lutein Treatment Effects on the Redox Status and Metalloproteinase-9 (MMP-9) in Oral Cancer Squamous Cells—Are There Therapeutical Hopes? 1 2,3 4 1 Dan Alexandru Enăs, escu , Mihaela Georgeta Moisescu , Marina Imre , Maria Greabu , Alexandra Ripszky Totan 1,* , Iulia Stanescu-Spinu 1, Marian Burcea 5, Crenguta Albu 6,* and Daniela Miricescu 1 1 Department of Biochemistry, Faculty of Dental Medicine, University of Medicine and Pharmacy Carol Davila, 8 Eroii Sanitari Blvd., Sector 5, 050474 Bucharest, Romania; [email protected] (D.A.E.); [email protected] (M.G.); [email protected] (I.S.-S.); [email protected] (D.M.) 2 Department Biophysics and Cellular Biotechnology, University of Medicine and Pharmacy Carol Davila, 8 Eroii Sanitari Blvd., Sector 5, 050474 Bucharest, Romania; [email protected] 3 Excellence Centre for Research in Biophysics and Cellular Biotechnology, University of Medicine and Pharmacy Carol Davila, 8 Eroii Sanitari Blvd., Sector 5, 050474 Bucharest, Romania 4 Department of Complete Denture, Faculty of Dental Medicine, Carol Davila University of Medicine and Pharmacy, 8 Eroii Sanitari Blvd., Sector 5, 050474 Bucharest, Romania; [email protected] 5 Department of Ophthalmology, Faculty of General Medicine, University of Medicine and Pharmacy Carol Davila, 8 Eroilor Sanitari Blvd., 050474 Bucharest, Romania; [email protected] 6 Department of Genetics, Faculty of General Medicine, University of Medicine and Pharmacy Carol Davila, 8 Eroilor Sanitari Blvd., 050474 Bucharest, Romania * Correspondence: [email protected] (A.R.T.); [email protected] (C.A.) Citation: En˘as, escu, D.A.; Moisescu, M.G.; Imre, M.; Greabu, M.; Ripszky Totan, A.; Stanescu-Spinu, I.; Burcea, Abstract: Carotenoids loaded in nanoparticles should be regarded as a promising way to increase M.; Albu, C.; Miricescu, D. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

(ER) Membrane Contact Sites (MCS) Uses Toxic Waste to Deliver Messages Edgar Djaha Yoboue1, Roberto Sitia1 and Thomas Simmen2
Yoboue et al. Cell Death and Disease (2018) 9:331 DOI 10.1038/s41419-017-0033-4 Cell Death & Disease REVIEW ARTICLE Open Access Redox crosstalk at endoplasmic reticulum (ER) membrane contact sites (MCS) uses toxic waste to deliver messages Edgar Djaha Yoboue1, Roberto Sitia1 and Thomas Simmen2 Abstract Many cellular redox reactions housed within mitochondria, peroxisomes and the endoplasmic reticulum (ER) generate hydrogen peroxide (H2O2) and other reactive oxygen species (ROS). The contribution of each organelle to the total cellular ROS production is considerable, but varies between cell types and also over time. Redox-regulatory enzymes are thought to assemble at a “redox triangle” formed by mitochondria, peroxisomes and the ER, assembling “redoxosomes” that sense ROS accumulations and redox imbalances. The redoxosome enzymes use ROS, potentially toxic by-products made by some redoxosome members themselves, to transmit inter-compartmental signals via chemical modifications of downstream proteins and lipids. Interestingly, important components of the redoxosome are ER chaperones and oxidoreductases, identifying ER oxidative protein folding as a key ROS producer and controller of the tri-organellar membrane contact sites (MCS) formed at the redox triangle. At these MCS, ROS accumulations could directly facilitate inter-organellar signal transmission, using ROS transporters. In addition, ROS influence the flux 2+ 2+ of Ca ions, since many Ca handling proteins, including inositol 1,4,5 trisphosphate receptors (IP3Rs), SERCA pumps or regulators of the mitochondrial Ca2+ uniporter (MCU) are redox-sensitive. Fine-tuning of these redox and ion signaling pathways might be difficult in older organisms, suggesting a dysfunctional redox triangle may accompany 1234567890 1234567890 the aging process. -

Deciphering the Gene Expression Profile of Peroxisome Proliferator
Chen et al. J Transl Med (2016) 14:157 DOI 10.1186/s12967-016-0871-3 Journal of Translational Medicine RESEARCH Open Access Deciphering the gene expression profile of peroxisome proliferator‑activated receptor signaling pathway in the left atria of patients with mitral regurgitation Mien‑Cheng Chen1*, Jen‑Ping Chang2, Yu‑Sheng Lin3, Kuo‑Li Pan3, Wan‑Chun Ho1, Wen‑Hao Liu1, Tzu‑Hao Chang4, Yao‑Kuang Huang5, Chih‑Yuan Fang1 and Chien‑Jen Chen1 Abstract Background: Differentially expressed genes in the left atria of mitral regurgitation (MR) pigs have been linked to peroxisome proliferator-activated receptor (PPAR) signaling pathway in the KEGG pathway. However, specific genes of the PPAR signaling pathway in the left atria of MR patients have never been explored. Methods: This study enrolled 15 MR patients with heart failure, 7 patients with aortic valve disease and heart failure, and 6 normal controls. We used PCR assay (84 genes) for PPAR pathway and quantitative RT-PCR to study specific genes of the PPAR pathway in the left atria. Results: Gene expression profiling analysis through PCR assay identified 23 genes to be differentially expressed in the left atria of MR patients compared to normal controls. The expressions of APOA1, ACADM, FABP3, ETFDH, ECH1, CPT1B, CPT2, SLC27A6, ACAA2, SMARCD3, SORBS1, EHHADH, SLC27A1, PPARGC1B, PPARA and CPT1A were significantly up-regulated, whereas the expression of PLTP was significantly down-regulated in the MR patients compared to normal controls. The expressions of HMGCS2, ACADM, FABP3, MLYCD, ECH1, ACAA2, EHHADH, CPT1A and PLTP were significantly up-regulated in the MR patients compared to patients with aortic valve disease. -

Follicular Thyroglobulin Enhances Gene Expression Necessary for Thyroid Hormone Secretion
Endocrine Journal 2015, 62 (11), 1007-1015 Original Follicular thyroglobulin enhances gene expression necessary for thyroid hormone secretion Yuko Ishido1), 2), 3), Yuqian Luo1), 3), Aya Yoshihara1), 3), Moyuru Hayashi3), Akio Yoshida2), Ichiro Hisatome2) and Koichi Suzuki1), 3) 1) Laboratory of Molecular Diagnostics, Department of Mycobacteriology, Leprosy Research Center, National Institute of Infectious Disease, Tokyo 189-0002, Japan 2) Division of Regenerative Medicine and Therapeutics, Department of Genetic Medicine and Regenerative Therapeutics, Institute of Regenerative Medicine and Biofunction, Tottori University Graduate School of Medical Science, Yonago, 683-8503, Japan 3) Department of Clinical Laboratory Science, Faculty of Medical Technology, Teikyo University, Tokyo 173-8605, Japan Abstract. We have previously shown that follicular thyroglobulin (Tg) has an unexpected function as an autocrine negative- feedback regulator of thyroid hormone (TH) biosynthesis. Tg significantly suppressed the expression of genes necessary for iodide transport and TH synthesis by counteracting stimulation by TSH. However, whether follicular Tg also regulates intracellular TH transport and its secretion from thyrocytes is not known. In the present study, we examined the potential effect of follicular Tg on TH transport and secretion by quantifying the expression of two TH transporters: monocarboxylate transporter 8 (MCT8) and μ-crystallin (CRYM). Our results showed that follicular Tg at physiologic concentrations enhanced both the mRNA and protein expression levels of MCT8 and CRYM in a time- and dose-dependent manner in rat thyroid FRTL-5 cells. Although both the sodium/iodide symporter (NIS), an essential transporter of iodide from blood into the thyroid, and MCT8, a transporter of synthesized TH from the gland, were co-localized on the basolateral membrane of rat thyrocytes in vivo, Tg decreased NIS expression and increased the expression of MCT8 by counteracting TSH action. -

Natural Course of Congenital Hypothyroidism by Dual Oxidase 2 Mutations from the Neonatal Period Through Puberty
Y Maruo and others Clinical features of DUOX2 174:4 453–463 Clinical Study defects Natural course of congenital hypothyroidism by dual oxidase 2 mutations from the neonatal period through puberty Yoshihiro Maruo1, Keisuke Nagasaki, Katsuyuki Matsui, Yu Mimura, Asami Mori, Maki Fukami2 and Yoshihiro Takeuchi Correspondence should be addressed Department of Pediatrics, Shiga University of Medical Science, Tsukinowa, Seta, Otsu, Shiga 520-2192, Japan, to Y Maruo 1Department of Pediatrics, Niigata University, Niigata, Japan and 2Department of Molecular Endocrinology, Email National Research Institute for Child Health and Development, Tokyo, Japan [email protected] Abstract Aim: We previously reported that biallelic mutations in dual oxidase 2 (DUOX2) cause transient hypothyroidism. Since then, many cases with DUOX2 mutations have been reported. However, the clinical features and prognosis of individuals with DUOX2 defects have not been clarified. Objective: We investigated the prognosis of patients with congenital hypothyroidism (CH) due to DUOX2 mutations. Patients: Twenty-five patients were identified by a neonatal screening program and included seven familial cases. Their serum TSH values ranged from 18.9 to 734.6 mU/l. Twenty-two of the patients had low serum free thyroxine (fT4) levels (0.17–1.1 ng/dl). Twenty-four of the patients were treated with L-thyroxine. Methods: We analyzed the DUOX2, thyroid peroxidase, NaC/IK symporter, and dual oxidase maturation factor 2 genes of these 25 patients by PCR-amplified direct sequencing. An additional 11 genes were analyzed in 11 of the 25 patients using next-generation sequencing. Results: All patients had biallelic DUOX2 mutations, and seven novel alleles were detected. -

Dissecting the Transcriptional Phenotype of Ribosomal Protein Deficiency: Implications for Diamond-Blackfan Anemia
Gene 545 (2014) 282–289 Contents lists available at ScienceDirect Gene journal homepage: www.elsevier.com/locate/gene Short Communication Dissecting the transcriptional phenotype of ribosomal protein deficiency: implications for Diamond-Blackfan Anemia Anna Aspesi a, Elisa Pavesi a, Elisa Robotti b, Rossella Crescitelli a,IleniaBoriac, Federica Avondo a, Hélène Moniz d, Lydie Da Costa d, Narla Mohandas e,PaolaRoncagliaf, Ugo Ramenghi g, Antonella Ronchi h, Stefano Gustincich f,SimoneMerlina,EmilioMarengob, Steven R. Ellis i, Antonia Follenzi a, Claudio Santoro a, Irma Dianzani a,⁎ a Department of Health Sciences, University of Eastern Piedmont, Novara, Italy b Department of Sciences and Technological Innovation, University of Eastern Piedmont, Alessandria, Italy c Department of Chemistry, University of Milan, Italy d U1009, AP-HP, Service d'Hématologie Biologique, Hôpital Robert Debré, Université Paris VII-Denis Diderot, Sorbonne Paris Cité, F-75475 Paris, France e New York Blood Center, NY, USA f International School for Advanced Studies (SISSA/ISAS), Trieste, Italy g Department of Pediatric Sciences, University of Torino, Torino, Italy h Department of Biotechnologies and Biosciences, Milano-Bicocca University, Italy i University of Louisville, KY, USA article info abstract Article history: Defects in genes encoding ribosomal proteins cause Diamond Blackfan Anemia (DBA), a red cell aplasia often as- Received 3 December 2013 sociated with physical abnormalities. Other bone marrow failure syndromes have been attributed to defects in Received in revised form 4 April 2014 ribosomal components but the link between erythropoiesis and the ribosome remains to be fully defined. Several Accepted 29 April 2014 lines of evidence suggest that defects in ribosome synthesis lead to “ribosomal stress” with p53 activation and Available online 15 May 2014 either cell cycle arrest or induction of apoptosis. -
When the Reaction Is
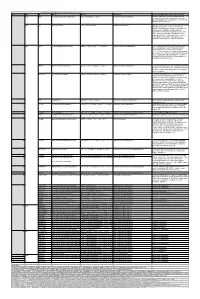
Table S3. iJL1678-ME model modification (blocked reactions) Iter. Cat. ID Name Formula Subsystem Comments (When the reaction is turned on) 1 bp2 EDD 6-phosphogluconate dehydratase 6pgc_c⇌2ddg6p_c + h2o_c Pentose Phosphate Pathway Create a major effect of steep acetate overflow elevation in high growth. Comparing to the main glycolytic pathway, it is metabolicly less efficient but proteomicly more efficient. bp1 ICL Isocitrate lyase icit_c→glx_c + succ_c Anaplerotic Reactions Bypass for the main TCA cycle pathways from turning isocitrate to succinate, when ICL is turned on, Isocitrate dehydrogenase(ICDHyr), 2-Oxogluterate dehydrogenase(AKGDH) and Succinyl-CoA synthetase (ATP-forming,SUCOAS) would reduce. Ref. (1) and (2) shows that this reaction is off in higher growth. Ref. (3) shows that this reaction is converging to being off when the dynamic of respiration using enzyme kinetics is simulated. 2 bp1 ABTA 4-aminobutyrate transaminase 4abut_c + akg_c⇌glu__L_c + sucsal_c Arginine and Proline Metabolism Another backup pathway of succinate production, from 2-Oxoglutarate (akg). Respiration would be induced when it is on, since the flux through ETC(CYTBO3_4pp and ATPS4rpp) would increase. As it requires the co-factor pyridoxal 5'-phosphate(2−) to get catalyzed(4), indicating that this reaction is regulated by the flux of other reactions(pyridoxal 5'- phosphate(2-) production, etc.). 3 GLYAT Glycine C-acetyltransferase accoa_c + gly_c⇌2aobut_c + coa_c Glycine and Serine Metabolism A reaction that back up for the respiration. Reactions fluxes in TCA cycle would drop when this reaction is turned on. It also requires pyridoxal 5'-phosphate(2−) for the regulation. 4 NADTRHD NAD transhydrogenase nad_c + nadph_c⇌nadh_c + nadp_c Oxidative Phosphorylation A reaction that would make the transition between NAD and NADP metabolically more efficient. -

Lapatinib Activates the Kelch-Like ECH- Associated Protein 1-Nuclear Factor Erythroid 2-Related Factor 2 Pathway in Hepg2 Cells
Updates in Pharmacology Book Chapter Lapatinib Activates the Kelch-Like ECH- Associated Protein 1-Nuclear Factor Erythroid 2-Related Factor 2 Pathway in HepG2 Cells Noëmi Johanna Roos1,2,3, Diell Aliu1,2, Jamal Bouitbir1,2,3 and Stephan Krähenbühl1,2,3* 1Division of Clinical Pharmacology & Toxicology, University Hospital, Switzerland 2Department of Biomedicine, University of Basel, Switzerland 3Swiss Centre for Applied Human Toxicology (SCAHT), Switzerland *Corresponding Author: Stephan Krähenbühl, Division of Clinical Pharmacology & Toxicology, University Hospital, Basel, Switzerland Published October 26, 2020 This Book Chapter is a republication of an article published by Stephan Krähenbühl, et al. at Frontiers in Pharmacology in June 2020. (Roos NJ, Aliu D, Bouitbir J and Krähenbühl S (2020) Lapatinib Activates the Kelch-Like ECH-Associated Protein 1- Nuclear Factor Erythroid 2-Related Factor 2 Pathway in HepG2 Cells. Front. Pharmacol. 11:944. doi: 10.3389/fphar.2020.00944) How to cite this book chapter: Noëmi Johanna Roos, Diell Aliu, Jamal Bouitbir, Stephan Krähenbühl. Lapatinib Activates the Kelch-Like ECH-Associated Protein 1-Nuclear Factor Erythroid 2-Related Factor 2 Pathway in HepG2 Cells. In: Nosheen Akhtar, editor. Updates in Pharmacology. Hyderabad, India: Vide Leaf. 2020. © The Author(s) 2020. This article is distributed under the terms of the Creative Commons Attribution 4.0 International 1 www.videleaf.com Updates in Pharmacology License(http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Data Availability Statement: All datasets for this study are included in the figshare repository: https://doi.org/10.6084/m9.figshare.12034608.v1. -

Independent Evolution of Four Heme Peroxidase Superfamilies
Archives of Biochemistry and Biophysics xxx (2015) xxx–xxx Contents lists available at ScienceDirect Archives of Biochemistry and Biophysics journal homepage: www.elsevier.com/locate/yabbi Independent evolution of four heme peroxidase superfamilies ⇑ Marcel Zámocky´ a,b, , Stefan Hofbauer a,c, Irene Schaffner a, Bernhard Gasselhuber a, Andrea Nicolussi a, Monika Soudi a, Katharina F. Pirker a, Paul G. Furtmüller a, Christian Obinger a a Department of Chemistry, Division of Biochemistry, VIBT – Vienna Institute of BioTechnology, University of Natural Resources and Life Sciences, Muthgasse 18, A-1190 Vienna, Austria b Institute of Molecular Biology, Slovak Academy of Sciences, Dúbravská cesta 21, SK-84551 Bratislava, Slovakia c Department for Structural and Computational Biology, Max F. Perutz Laboratories, University of Vienna, A-1030 Vienna, Austria article info abstract Article history: Four heme peroxidase superfamilies (peroxidase–catalase, peroxidase–cyclooxygenase, peroxidase–chlo- Received 26 November 2014 rite dismutase and peroxidase–peroxygenase superfamily) arose independently during evolution, which and in revised form 23 December 2014 differ in overall fold, active site architecture and enzymatic activities. The redox cofactor is heme b or Available online xxxx posttranslationally modified heme that is ligated by either histidine or cysteine. Heme peroxidases are found in all kingdoms of life and typically catalyze the one- and two-electron oxidation of a myriad of Keywords: organic and inorganic substrates. In addition to this peroxidatic activity distinct (sub)families show pro- Heme peroxidase nounced catalase, cyclooxygenase, chlorite dismutase or peroxygenase activities. Here we describe the Peroxidase–catalase superfamily phylogeny of these four superfamilies and present the most important sequence signatures and active Peroxidase–cyclooxygenase superfamily Peroxidase–chlorite dismutase superfamily site architectures.