2.2.14 Viral Encephalopathy and Retinopathy
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ABSTRACT LUCKENBACH, JOHN ADAM. Breeding Biotechnology
ABSTRACT LUCKENBACH, JOHN ADAM. Breeding Biotechnology, Sex Determination, and Growth in Southern flounder, Paralichthys lethostigma. (Under the direction of Dr. John Godwin and Dr. Russell J. Borski). Southern flounder (Paralichthys lethostigma) support valuable, but declining US fisheries. This species is therefore a strong candidate for aquaculture to mitigate fishing impacts and stabilize seafood supply. Because female flounder reach substantially larger sizes than males, all-female culture is desirable for commercial aquaculture. Hence, a thorough understanding of sexual development, its timing and regulation by temperature is essential for optimization of flounder aquaculture. To better understand ovarian and testicular development in southern flounder, structural and cellular sex-distinguishing markers were studied using histological methods. We found that histologically discernible sex differentiation occurs in southern flounder at ~75-120 mm TL and that early differentiation is considerably delayed relative to its Japanese congener, P. olivaceus. High (28°C) and low (18°C) water temperatures, produced a higher proportion of males (96% and 78% males, respectively). The sex ratio at a mid-range (23°C) temperature was not different from 1:1. This suggests that southern flounder possess a temperature sensitive mechanism of sex determination. Growth was also affected by temperature with the temperature that maximized females inducing better growth. Aromatase cytochrome P450 (P450arom) is responsible for estrogen biosynthesis and plays a critical role in ovarian differentiation. We cloned ovarian P450arom and developed a qRT-PCR for assessment of early sex differentiation. The deduced amino acid sequence for southern flounder P450arom is very similar to P450arom in other teleosts. Comparison of P450arom intron sequences of southern flounder within and between different populations revealed substantial inter-individual variation that may affect sex determination responses. -
Schedule Onlinepdf
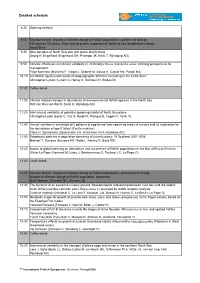
Detailed schedule 8:30 Opening session 9:00 Keynote lecture: Impacts of climate change on flatfish populations - patterns of change 100 days to 100 years: Short and long-term responses of flatfish to sea temperature change David Sims 9:30 Nine decades of North Sea sole and plaice distributions Georg H. Engelhard (Engelhard GH, Pinnegar JK, Kell LT, Rijnsdorp AD) 9:50 Climatic effects on recruitment variability in Platichthys flesus and Solea solea: defining perspectives for management. Filipe Martinho (Martinho F, Viegas I, Dolbeth M, Sousa H, Cabral HN, Pardal MA) 10:10 Are flatfish species with southern biogeographic affinities increasing in the Celtic Sea? Christopher Lynam (Lynam C, Harlay X, Gerritsen H, Stokes D) 10:30 Coffee break 11:00 Climate related changes in abundance of non-commercial flatfish species in the North Sea Ralf van Hal (van Hal R, Smits K, Rijnsdorp AD) 11:20 Inter-annual variability of potential spawning habitat of North Sea plaice Christophe Loots (Loots C, Vaz S, Koubii P, Planque B, Coppin F, Verin Y) 11:40 Annual variation in simulated drift patterns of egg/larvae from spawning areas to nursery and its implication for the abundance of age-0 turbot (Psetta maxima) Claus R. Sparrevohn (Sparrevohn CR, Hinrichsen H-H, Rijnsdorp AD) 12:00 Broadscale patterns in population dynamics of juvenile plaice: W Scotland 2001-2008 Michael T. Burrows (Burrows MT, Robb L, Harvey R, Batty RS) 12:20 Impact of global warming on abundance and occurrence of flatfish populations in the Bay of Biscay (France) Olivier Le Pape (Hermant -

229 Index of Scientific and Vernacular Names
previous page 229 INDEX OF SCIENTIFIC AND VERNACULAR NAMES EXPLANATION OF THE SYSTEM Type faces used: Italics : Valid scientific names (genera and species) Italics : Synonyms * Italics : Misidentifications (preceded by an asterisk) ROMAN (saps) : Family names Roman : International (FAO) names of species 230 Page Page A African red snapper ................................................. 79 Abalistes stellatus ............................................... 42 African sawtail catshark ......................................... 144 Abámbolo ............................................................... 81 African sicklefìsh ...................................................... 62 Abámbolo de bajura ................................................ 81 African solenette .................................................... 111 Ablennes hians ..................................................... 44 African spadefish ..................................................... 63 Abuete cajeta ........................................................ 184 African spider shrimp ............................................. 175 Abuete de Angola ................................................. 184 African spoon-nose eel ............................................ 88 Abuete negro ........................................................ 184 African squid .......................................................... 199 Abuete real ........................................................... 183 African striped grunt ................................................ -

Consequences of Hatchery Stocking
bioRxiv preprint doi: https://doi.org/10.1101/828798; this version posted November 2, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Consequences of hatchery stocking: 2 Lessons from Japan 3 4 Sort title: Japan hatchery stocking 5 6 Shuichi Kitada 7 8 1Tokyo University of Marine Science and Technology, Tokyo 108-8477, Japan. 9 E-mail: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/828798; this version posted November 2, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 10 Abstract 11 More than 26 billion juveniles of 180 marine species are released annually into the wild in 12 over 20 countries, but the usefulness remains unclear. Here, I review the effects of stocking by 13 Japanese marine and salmon stock-enhancement programmes, and evaluate the efficacy 14 through a meta-analysis, with new cases added to those previously considered. The posterior 15 mean recapture rates (±SD) was 8.2 ± 4.7%; mean economic efficiency was 2.7 ± 4.5, with 16 many cases having values of 1–2, excluding consideration for personnel costs and negative 17 impacts on wild populations. At the macro-scale, the proportion of released seeds to total 18 catch was 76 ± 20% for Japanese scallop, 28 ± 10% for abalone, 20 ± 5% for swimming crab, 19 13 ± 5% for kuruma prawn, 11 ± 4% for Japanese flounder and 7 ± 2% for red sea bream. -

Session Summaries-2008
Session Summaries-2008 SUMMARY OF SCIENTIFIC SESSIONS AND WORKSHOPS Science Board Symposium (S1) Beyond observations to achieving understanding and forecasting in a changing North Pacific: Forward to the FUTURE Co-Convenors: John E. Stein (SB), Michael J. Dagg (BIO), Gordon H. Kruse (FIS), Glen S. Jamieson (MEQ), Hiroya Sugisaki (MONITOR), Michael G. Foreman (POC), Bernard A. Megrey (TCODE), Harold P. Batchelder (CCCC), Michio J. Kishi (CCCC), Fangli Qiao (China), Sinjae Yoo (Korea) and Mikhail Stepanenko (Russia) Background FUTURE (Forecasting and Understanding Trends, Uncertainty and Responses of North Pacific Marine Ecosystems), the new Science Program undertaken by PICES member countries, has the broad goals of: (1) understanding the responses of marine ecosystems in the North Pacific to climate change and human activities at basin-wide and regional scales; (2) providing forecasts of what might be expected based on a current understanding of how nature works; and (3) communicating this information effectively to its members and to society in general. Past advances in understanding marine ecosystems in the North Pacific have been largely based either on the direct analysis of observations, or on the development of conceptual and numerical models that help to describe the processes underlying the observations. Though these activities will continue to play an important role in FUTURE, the provision of forecasts and estimates of their associated uncertainties necessitates moving beyond observationally based understanding, so that ecosystem responses to natural and anthropogenic changes can be anticipated and communicated effectively to society. Presentations were invited to address the goals of FUTURE and the three key research questions that it identifies: 1. -

Shellfish Diseases and Their Management in Commercial Recirculating Systems
Shellfish Diseases and Their Management in Commercial Recirculating Systems Ralph Elston AquaTechnics & Pacific Shellfish Institute PO Box 687 Carlsborg, WA 98324 Introduction Intensive culture of early life stages of bivalve shellfish culture has been practiced since at least the late 1950’s on an experimental basis. Production scale culture emerged in the 1970’s and today, hathcheries and nurseries produce large numbers of a variety of species of oysters, clams and scallops. The early life stage systems may be entirely or partially recirculating or static. Management of infectious diseases in these systems has been a challenge since their inception and effective health management is a requisite to successful culture. The diseases which affect early life stage shellfish in intensive production systems and the principles and practice of health management are the subject of this presentation. Shellfish Diseases and Management Diseases of bivalve shellfish affecting those reared or harvested from extensive culture primarily consist of parasitic infections and generally comprise the reportable or certifiable diseases. Due to the extensive nature of such culture, intervention options or disease control are limited. In contrast, infectious diseases known from early life stages in intensive culture systems tend to be opportunistic in nature and offer substantial opportunity for management due to the control that can be exerted at key points in the systems. In marine shellfish hatcheries, infectious organisms can enter the system from three sources: brood stock, seawater source and algal food source. Once an organism is established in the system, it may persist without further introduction. Bacterial infections are the most common opportunistic infection in shellfish hatcheries. -
2018 Final LOFF W/ Ref and Detailed Info
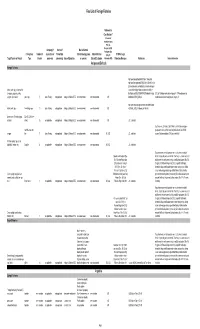
Final List of Foreign Fisheries Rationale for Classification ** (Presence of mortality or injury (P/A), Co- Occurrence (C/O), Company (if Source of Marine Mammal Analogous Gear Fishery/Gear Number of aquaculture or Product (for Interactions (by group Marine Mammal (A/G), No RFMO or Legal Target Species or Product Type Vessels processor) processing) Area of Operation or species) Bycatch Estimates Information (N/I)) Protection Measures References Detailed Information Antigua and Barbuda Exempt Fisheries http://www.fao.org/fi/oldsite/FCP/en/ATG/body.htm http://www.fao.org/docrep/006/y5402e/y5402e06.htm,ht tp://www.tradeboss.com/default.cgi/action/viewcompan lobster, rock, spiny, demersal fish ies/searchterm/spiny+lobster/searchtermcondition/1/ , (snappers, groupers, grunts, ftp://ftp.fao.org/fi/DOCUMENT/IPOAS/national/Antigua U.S. LoF Caribbean spiny lobster trap/ pot >197 None documented, surgeonfish), flounder pots, traps 74 Lewis Fishing not applicable Antigua & Barbuda EEZ none documented none documented A/G AndBarbuda/NPOA_IUU.pdf Caribbean mixed species trap/pot are category III http://www.nmfs.noaa.gov/pr/interactions/fisheries/tabl lobster, rock, spiny free diving, loops 19 Lewis Fishing not applicable Antigua & Barbuda EEZ none documented none documented A/G e2/Atlantic_GOM_Caribbean_shellfish.html Queen conch (Strombus gigas), Dive (SCUBA & free molluscs diving) 25 not applicable not applicable Antigua & Barbuda EEZ none documented none documented A/G U.S. trade data Southeastern U.S. Atlantic, Gulf of Mexico, and Caribbean snapper- handline, hook and grouper and other reef fish bottom longline/hook-and-line/ >5,000 snapper line 71 Lewis Fishing not applicable Antigua & Barbuda EEZ none documented none documented N/I, A/G U.S. -

Deep-Bottom Fish Identification Cards for Small-Scale Fishermen
Deep-bottom fish identification cards for small-scale fishermen iq © Secretariat of the Pacific Community, 2013 These identification cards are produced by the Secretariat of the Pacific Community (SPC) to help improve catch data and statistics from small-scale vessels that regularly target deep-bottom fish species. With a better understanding of small-scale vessel catches, regional fisheries managers can ensure that stocks are better managed throughout their range and that small-scale fishermen get more recognition and support for their contribution to food security and employment at the local level. The most likely users of these cards are small-scale fishers who actively target deep-bottom fish stocks, along with fishery officers who are tasked with monitoring these fisheries. Fisheries training institutions are other potential users. Printing of these cards was made possible through financial assistance provided by AusAID. Project coordination was done by the Oceanic Fisheries Programme’s Ecosystem Monitoring Section and the Fisheries Information Section, SPC. Illustrations were made by Les Hata and Rachel O’Shea. This publication is based on the Fish species identification manual for deep-bottom snapper fishermen by Chapman et al. (2008). Secretariat of the Pacific Community Cataloguing-in-publication data Deep-bottom fish identification cards for small-scale fishermen / produced by the Secretariat of the Pacific Community 1. Marine fishes – Classification – Oceania. 2. Fishery management – Oceania. 3. Fishes – Identification – Oceania. I. Title II. Secretariat of the Pacific Community 597.0995 AACR2 ISBN: 978-982-00-0589-1 Head Body Caudal fin Different caudal fin types Spiny dorsal fin Soft dorsal fin Rounded Dorsal spines Opercular spine Truncate Snout Emarginate Lateral line Lunate Cheek Caudal fin Gill cover Forked Anal fin Cheek cleft Pectoral fin Pelvic fin Anal spines Snappers 2 Ruby snapper Etelis carbunculus Dorsal fin deeply notched Distinct yellowish Small black markings on lateral line upper lobe of caudal fin max. -

Smithsonian Miscellaneous Collections
SMITHSONIAN MISCELLANEOUS COLLECTIONS VOLUME 98, NUMBER 25 ShSan a* INSTyJJ am 4 LIST OF THE FISHES TAKEN ON THE PRESIDENTIAL CRUISE OF 19 &mi .mtfL BY WALDO L. SCHMITT AND LEONARD P. SGHULTZ U. S. National Museum (Publication 3551) CITY OF WASHINGTON PUBLISHED BY THE SMITHSONIAN INSTITUTION JANUARY 4, 1940 L SMITHSONIAN MISCELLANEOUS COLLECTIONS VOLUME 98, NUMBER 25 LIST OF THE FISHES TAKEN ON THE PRESIDENTIAL CRUISE OF 1938 BY WALDO L. SCHMITT AND LEONARD P. SCHULTZ U. S. National Museum ft&sss*?; %><asssSa 5«5 £9 >®g»£fc (Publication- 3551) CITY OF WASHINGTON PUBLISHED BY THE SMITHSONIAN INSTITUTION JANUARY 4, 1940 Zfye £ovb (gaftimore (pree* BALTIMORE!. MD.. IT. S. A. LIST OF THE FISHES TAKEN ON THE PRESIDENTIAL CRUISE OF 1938 By WALDO L. SCHMITT and LEONARD P. SCHULTZ U. S. National Museum This list comprises only those fishes collected on the Presidential Cruise of 1938 which have actually been incorporated in the study collections of the United States National Museum. President Roose- velt, himself an ardent fisherman and keen naturalist, was the principal contributor of the specimens listed. Not only did he obtain some rarities, but he also succeeded in establishing maximum weight records for two species : 20 pounds for the rainbow runner, Elagatis bipinmdatus, and 38 pounds for the blue crevally, Caranx stellatus. The President was most ably assisted by the members of his personal party : Frederick B. Adams ; Basil O'Connor ; Stephen T. Early, the President's Secretary ; Capt. Ross T. Mclntire, Medical Corps, U.S.N., Aide; Capt. D. J. Callaghan, U.S.N. , Naval Aide; and Col. -

Betanodavirus and VER Disease: a 30-Year Research Review
pathogens Review Betanodavirus and VER Disease: A 30-year Research Review Isabel Bandín * and Sandra Souto Departamento de Microbioloxía e Parasitoloxía-Instituto de Acuicultura, Universidade de Santiago de Compostela, 15782 Santiago de Compostela, Spain; [email protected] * Correspondence: [email protected] Received: 20 December 2019; Accepted: 4 February 2020; Published: 9 February 2020 Abstract: The outbreaks of viral encephalopathy and retinopathy (VER), caused by nervous necrosis virus (NNV), represent one of the main infectious threats for marine aquaculture worldwide. Since the first description of the disease at the end of the 1980s, a considerable amount of research has gone into understanding the mechanisms involved in fish infection, developing reliable diagnostic methods, and control measures, and several comprehensive reviews have been published to date. This review focuses on host–virus interaction and epidemiological aspects, comprising viral distribution and transmission as well as the continuously increasing host range (177 susceptible marine species and epizootic outbreaks reported in 62 of them), with special emphasis on genotypes and the effect of global warming on NNV infection, but also including the latest findings in the NNV life cycle and virulence as well as diagnostic methods and VER disease control. Keywords: nervous necrosis virus (NNV); viral encephalopathy and retinopathy (VER); virus–host interaction; epizootiology; diagnostics; control 1. Introduction Nervous necrosis virus (NNV) is the causative agent of viral encephalopathy and retinopathy (VER), otherwise known as viral nervous necrosis (VNN). The disease was first described at the end of the 1980s in Australia and in the Caribbean [1–3], and has since caused a great deal of mortalities and serious economic losses in a variety of reared marine fish species, but also in freshwater species worldwide. -

Proceedings of the 40Th U.S.-Japan Aquaculture Panel Symposium
Hatchery Technology for High Quality Juvenile Production Proceedings of the 40th U.S.-Japan Aquaculture Panel Symposium University of Hawaii East West Center Honolulu, Hawaii October 22-23 2012 U.S. DEPARTMENT OF COMMERCE National Oceanic and Atmospheric Administration National Marine Fisheries Service NOAA Technical Memorandum NMFS-F/SPO-136 Hatchery Technology for High Quality Juvenile Production Proceedings of the 40th U.S.-Japan Aquaculture Panel Symposium University of Hawaii East West Center Honolulu, Hawaii October 22-23 2012 Mike Rust1, Paul Olin2, April Bagwill3, and Marie Fujitani3, editors 1Northwest Fisheries Science Center 2725 Montlake Boulevard East Seattle, Washington 98112 2California Sea Grant UCSD / Scripps Institution of Oceanography 133 Aviation Blvd., Suite 109 Santa Rosa CA 95403 3NOAA National Marine Fisheries Service 1315 East-West Highway Silver Spring, MD 20910 NOAA Technical Memorandum NMFS-F/SPO-136 December 2013 U.S. Department of Commerce Penny Pritzker, Secretary of Commerce National Oceanic and Atmospheric Administration Dr. Kathryn Sullivan, (Acting) NOAA Administrator National Marine Fisheries Service Samuel D. Rauch III, (Acting) Assistant Administrator for Fisheries SUGGESTED CITATION: Rust, M., P. Olin, A. Bagwill and M. Fujitani (editors). 2013. Hatchery Technology for High Quality Juvenile Production: Proceedings of the 40th U.S.-Japan Aquaculture Panel Symposium, Honolulu, Hawaii, October 22-23, 2012. U.S. Dept. Commerce, NOAA Tech. Memo. NMFS-F/SPO-136. A COPY OF THIS REPORT MAY BE OBTAINED FROM: Northwest Fisheries Science Center 2725 Montlake Boulevard East Seattle, Washington 98112 OR ONLINE AT: http://spo.nmfs.noaa.gov/tm/ Reference throughout this document to trade names does not imply endorsement by the National Marine Fisheries Service, NOAA. -

Deep Bottom ID
by par Lindsay Chapman, Aymeric Desurmont, Youngmi Choi, Lindsay Chapman, Aymeric Desurmont, Youngmi Choi, Pierre Boblin, William Sokimi and Steve Beverly Pierre Boblin, William Sokimi et Steve Beverly nzaid ii © Copyright Secretariat of the Pacific Community 2008 © Copyright Secrétariat général de la Communauté du Pacifique 2008 All rights for commercial / for profit reproduction or translation, in any Tous droits réservés de reproduction ou de traduction à des fins commer- form, reserved. SPC authorises the partial reproduction or translation of ciales/lucratives, sous quelque forme que ce soit. Le Secrétariat général de this material for scientific, educational or research purposes, provided SPC la Communauté du Pacifique autorise la reproduction ou la traduction par- and the source document are properly acknowledged. Permission to tielles de ce document à des fins scientifiques ou éducatives ou pour les reproduce the document and/or translate in whole, in any form, whether besoins de la recherche, à condition qu’il soit fait mention de la CPS et de for commercial / for profit or non-profit purposes, must be requested in la source. L’autorisation de la reproduction et/ou de la traduction intégrale writing. Original SPC artwork may not be altered or separately published ou partielle de ce document, sous quelque forme que ce soit, à des fins without permission. commerciales/lucratives ou à titre gratuit, doit être sollicitée au préalable par écrit. Il est interdit de modifier ou de publier séparément des Original text: English and French graphismes originaux de la CPS sans autorisation préalable. Texte original :anglais et français Secretariat of the Pacific Community Cataloguing-in-publication data Chapman, L.