Microsatellite Analysis As a Tool for Discriminating an Interfamily Hybrid Between Olive Flounder and Starry Flounder
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ABSTRACT LUCKENBACH, JOHN ADAM. Breeding Biotechnology
ABSTRACT LUCKENBACH, JOHN ADAM. Breeding Biotechnology, Sex Determination, and Growth in Southern flounder, Paralichthys lethostigma. (Under the direction of Dr. John Godwin and Dr. Russell J. Borski). Southern flounder (Paralichthys lethostigma) support valuable, but declining US fisheries. This species is therefore a strong candidate for aquaculture to mitigate fishing impacts and stabilize seafood supply. Because female flounder reach substantially larger sizes than males, all-female culture is desirable for commercial aquaculture. Hence, a thorough understanding of sexual development, its timing and regulation by temperature is essential for optimization of flounder aquaculture. To better understand ovarian and testicular development in southern flounder, structural and cellular sex-distinguishing markers were studied using histological methods. We found that histologically discernible sex differentiation occurs in southern flounder at ~75-120 mm TL and that early differentiation is considerably delayed relative to its Japanese congener, P. olivaceus. High (28°C) and low (18°C) water temperatures, produced a higher proportion of males (96% and 78% males, respectively). The sex ratio at a mid-range (23°C) temperature was not different from 1:1. This suggests that southern flounder possess a temperature sensitive mechanism of sex determination. Growth was also affected by temperature with the temperature that maximized females inducing better growth. Aromatase cytochrome P450 (P450arom) is responsible for estrogen biosynthesis and plays a critical role in ovarian differentiation. We cloned ovarian P450arom and developed a qRT-PCR for assessment of early sex differentiation. The deduced amino acid sequence for southern flounder P450arom is very similar to P450arom in other teleosts. Comparison of P450arom intron sequences of southern flounder within and between different populations revealed substantial inter-individual variation that may affect sex determination responses. -
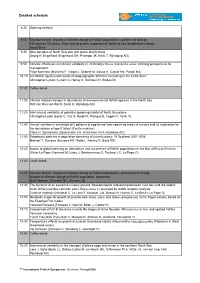
Schedule Onlinepdf
Detailed schedule 8:30 Opening session 9:00 Keynote lecture: Impacts of climate change on flatfish populations - patterns of change 100 days to 100 years: Short and long-term responses of flatfish to sea temperature change David Sims 9:30 Nine decades of North Sea sole and plaice distributions Georg H. Engelhard (Engelhard GH, Pinnegar JK, Kell LT, Rijnsdorp AD) 9:50 Climatic effects on recruitment variability in Platichthys flesus and Solea solea: defining perspectives for management. Filipe Martinho (Martinho F, Viegas I, Dolbeth M, Sousa H, Cabral HN, Pardal MA) 10:10 Are flatfish species with southern biogeographic affinities increasing in the Celtic Sea? Christopher Lynam (Lynam C, Harlay X, Gerritsen H, Stokes D) 10:30 Coffee break 11:00 Climate related changes in abundance of non-commercial flatfish species in the North Sea Ralf van Hal (van Hal R, Smits K, Rijnsdorp AD) 11:20 Inter-annual variability of potential spawning habitat of North Sea plaice Christophe Loots (Loots C, Vaz S, Koubii P, Planque B, Coppin F, Verin Y) 11:40 Annual variation in simulated drift patterns of egg/larvae from spawning areas to nursery and its implication for the abundance of age-0 turbot (Psetta maxima) Claus R. Sparrevohn (Sparrevohn CR, Hinrichsen H-H, Rijnsdorp AD) 12:00 Broadscale patterns in population dynamics of juvenile plaice: W Scotland 2001-2008 Michael T. Burrows (Burrows MT, Robb L, Harvey R, Batty RS) 12:20 Impact of global warming on abundance and occurrence of flatfish populations in the Bay of Biscay (France) Olivier Le Pape (Hermant -

The Osmoregulatory Metabolism Op the Starry Flounder, Platichthys Stellatus
THE OSMOREGULATORY METABOLISM OP THE STARRY FLOUNDER, PLATICHTHYS STELLATUS by CLEVELAND PENDLETON HICKMAN, JR. B.A., DePauw University, 1950 M.S., University of New Hampshire, 1953 A THESIS SUBMITTED IN PARTIAL FULFILMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY in the Department of Zoology We accept this thesis as conforming to the required standard. THE UNIVERSITY OF BRITISH COLUMBIA June, 1958 Faculty of Graduate Studies PROGRAMME OF THE FINAL ORAL EXAMINATION FOR THE DEGREE OF DOCTOR OF PHILOSOPHY of CLEVELAND PENDLETON HICKMAN JR. B.A. DePauw University, 1950 M.S. University of New Hampshire, 1953 IN ROOM 187A, BIOLOGICAL SCIENCES BUILDING MONDAY, JUNE 30, 1958 at 10:30 a.m. COMMITTEE IN CHARGE DEAN F. H. SOWARD, Chairman H. ADASKIN W. S. HOAR W. A. CLEMENS W. N. HOLMES I. McT. COWAN C. C. LINDSEY P. A. DEHNEL H. McLENNAN R. F. SCAGEL External Examiner: F. E. J. FRY University of Toronto THE OSMOREGULATORY METABOLISM OF THE STARRY FLOUNDER, PLATICHTYS STELLATUS ABSTRACT Energy demands for osmotic regulation and the possible osmoregulatory role of the thyroid gland were investigated in the euryhaline starry flounder, Platichthys stellatus. Using a melt• ing-point technique, it was established that flounder could regulate body fluid concentration independent of widely divergent environ• mental salinities. Small flounder experienced more rapid disturb• ances of body fluid concentration than large flounder after abrupt salinity alterations. The standard metabolic rate of flounder adapted to fresh water was consistently and significantly less than that of marine flounder. In supernormal salinities standard metabolic rate was significantly greater than in normal sea water. -

Preliminary Mass-Balance Food Web Model of the Eastern Chukchi Sea
NOAA Technical Memorandum NMFS-AFSC-262 Preliminary Mass-balance Food Web Model of the Eastern Chukchi Sea by G. A. Whitehouse U.S. DEPARTMENT OF COMMERCE National Oceanic and Atmospheric Administration National Marine Fisheries Service Alaska Fisheries Science Center December 2013 NOAA Technical Memorandum NMFS The National Marine Fisheries Service's Alaska Fisheries Science Center uses the NOAA Technical Memorandum series to issue informal scientific and technical publications when complete formal review and editorial processing are not appropriate or feasible. Documents within this series reflect sound professional work and may be referenced in the formal scientific and technical literature. The NMFS-AFSC Technical Memorandum series of the Alaska Fisheries Science Center continues the NMFS-F/NWC series established in 1970 by the Northwest Fisheries Center. The NMFS-NWFSC series is currently used by the Northwest Fisheries Science Center. This document should be cited as follows: Whitehouse, G. A. 2013. A preliminary mass-balance food web model of the eastern Chukchi Sea. U.S. Dep. Commer., NOAA Tech. Memo. NMFS-AFSC-262, 162 p. Reference in this document to trade names does not imply endorsement by the National Marine Fisheries Service, NOAA. NOAA Technical Memorandum NMFS-AFSC-262 Preliminary Mass-balance Food Web Model of the Eastern Chukchi Sea by G. A. Whitehouse1,2 1Alaska Fisheries Science Center 7600 Sand Point Way N.E. Seattle WA 98115 2Joint Institute for the Study of the Atmosphere and Ocean University of Washington Box 354925 Seattle WA 98195 www.afsc.noaa.gov U.S. DEPARTMENT OF COMMERCE Penny. S. Pritzker, Secretary National Oceanic and Atmospheric Administration Kathryn D. -

Consequences of Hatchery Stocking
bioRxiv preprint doi: https://doi.org/10.1101/828798; this version posted November 2, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Consequences of hatchery stocking: 2 Lessons from Japan 3 4 Sort title: Japan hatchery stocking 5 6 Shuichi Kitada 7 8 1Tokyo University of Marine Science and Technology, Tokyo 108-8477, Japan. 9 E-mail: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/828798; this version posted November 2, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 10 Abstract 11 More than 26 billion juveniles of 180 marine species are released annually into the wild in 12 over 20 countries, but the usefulness remains unclear. Here, I review the effects of stocking by 13 Japanese marine and salmon stock-enhancement programmes, and evaluate the efficacy 14 through a meta-analysis, with new cases added to those previously considered. The posterior 15 mean recapture rates (±SD) was 8.2 ± 4.7%; mean economic efficiency was 2.7 ± 4.5, with 16 many cases having values of 1–2, excluding consideration for personnel costs and negative 17 impacts on wild populations. At the macro-scale, the proportion of released seeds to total 18 catch was 76 ± 20% for Japanese scallop, 28 ± 10% for abalone, 20 ± 5% for swimming crab, 19 13 ± 5% for kuruma prawn, 11 ± 4% for Japanese flounder and 7 ± 2% for red sea bream. -

Session Summaries-2008
Session Summaries-2008 SUMMARY OF SCIENTIFIC SESSIONS AND WORKSHOPS Science Board Symposium (S1) Beyond observations to achieving understanding and forecasting in a changing North Pacific: Forward to the FUTURE Co-Convenors: John E. Stein (SB), Michael J. Dagg (BIO), Gordon H. Kruse (FIS), Glen S. Jamieson (MEQ), Hiroya Sugisaki (MONITOR), Michael G. Foreman (POC), Bernard A. Megrey (TCODE), Harold P. Batchelder (CCCC), Michio J. Kishi (CCCC), Fangli Qiao (China), Sinjae Yoo (Korea) and Mikhail Stepanenko (Russia) Background FUTURE (Forecasting and Understanding Trends, Uncertainty and Responses of North Pacific Marine Ecosystems), the new Science Program undertaken by PICES member countries, has the broad goals of: (1) understanding the responses of marine ecosystems in the North Pacific to climate change and human activities at basin-wide and regional scales; (2) providing forecasts of what might be expected based on a current understanding of how nature works; and (3) communicating this information effectively to its members and to society in general. Past advances in understanding marine ecosystems in the North Pacific have been largely based either on the direct analysis of observations, or on the development of conceptual and numerical models that help to describe the processes underlying the observations. Though these activities will continue to play an important role in FUTURE, the provision of forecasts and estimates of their associated uncertainties necessitates moving beyond observationally based understanding, so that ecosystem responses to natural and anthropogenic changes can be anticipated and communicated effectively to society. Presentations were invited to address the goals of FUTURE and the three key research questions that it identifies: 1. -

Shellfish Diseases and Their Management in Commercial Recirculating Systems
Shellfish Diseases and Their Management in Commercial Recirculating Systems Ralph Elston AquaTechnics & Pacific Shellfish Institute PO Box 687 Carlsborg, WA 98324 Introduction Intensive culture of early life stages of bivalve shellfish culture has been practiced since at least the late 1950’s on an experimental basis. Production scale culture emerged in the 1970’s and today, hathcheries and nurseries produce large numbers of a variety of species of oysters, clams and scallops. The early life stage systems may be entirely or partially recirculating or static. Management of infectious diseases in these systems has been a challenge since their inception and effective health management is a requisite to successful culture. The diseases which affect early life stage shellfish in intensive production systems and the principles and practice of health management are the subject of this presentation. Shellfish Diseases and Management Diseases of bivalve shellfish affecting those reared or harvested from extensive culture primarily consist of parasitic infections and generally comprise the reportable or certifiable diseases. Due to the extensive nature of such culture, intervention options or disease control are limited. In contrast, infectious diseases known from early life stages in intensive culture systems tend to be opportunistic in nature and offer substantial opportunity for management due to the control that can be exerted at key points in the systems. In marine shellfish hatcheries, infectious organisms can enter the system from three sources: brood stock, seawater source and algal food source. Once an organism is established in the system, it may persist without further introduction. Bacterial infections are the most common opportunistic infection in shellfish hatcheries. -

Humboldt Bay Fishes
Humboldt Bay Fishes ><((((º>`·._ .·´¯`·. _ .·´¯`·. ><((((º> ·´¯`·._.·´¯`·.. ><((((º>`·._ .·´¯`·. _ .·´¯`·. ><((((º> Acknowledgements The Humboldt Bay Harbor District would like to offer our sincere thanks and appreciation to the authors and photographers who have allowed us to use their work in this report. Photography and Illustrations We would like to thank the photographers and illustrators who have so graciously donated the use of their images for this publication. Andrey Dolgor Dan Gotshall Polar Research Institute of Marine Sea Challengers, Inc. Fisheries And Oceanography [email protected] [email protected] Michael Lanboeuf Milton Love [email protected] Marine Science Institute [email protected] Stephen Metherell Jacques Moreau [email protected] [email protected] Bernd Ueberschaer Clinton Bauder [email protected] [email protected] Fish descriptions contained in this report are from: Froese, R. and Pauly, D. Editors. 2003 FishBase. Worldwide Web electronic publication. http://www.fishbase.org/ 13 August 2003 Photographer Fish Photographer Bauder, Clinton wolf-eel Gotshall, Daniel W scalyhead sculpin Bauder, Clinton blackeye goby Gotshall, Daniel W speckled sanddab Bauder, Clinton spotted cusk-eel Gotshall, Daniel W. bocaccio Bauder, Clinton tube-snout Gotshall, Daniel W. brown rockfish Gotshall, Daniel W. yellowtail rockfish Flescher, Don american shad Gotshall, Daniel W. dover sole Flescher, Don stripped bass Gotshall, Daniel W. pacific sanddab Gotshall, Daniel W. kelp greenling Garcia-Franco, Mauricio louvar -
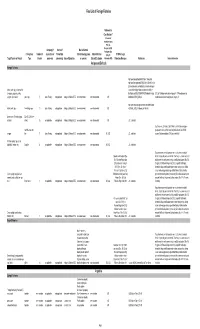
2018 Final LOFF W/ Ref and Detailed Info
Final List of Foreign Fisheries Rationale for Classification ** (Presence of mortality or injury (P/A), Co- Occurrence (C/O), Company (if Source of Marine Mammal Analogous Gear Fishery/Gear Number of aquaculture or Product (for Interactions (by group Marine Mammal (A/G), No RFMO or Legal Target Species or Product Type Vessels processor) processing) Area of Operation or species) Bycatch Estimates Information (N/I)) Protection Measures References Detailed Information Antigua and Barbuda Exempt Fisheries http://www.fao.org/fi/oldsite/FCP/en/ATG/body.htm http://www.fao.org/docrep/006/y5402e/y5402e06.htm,ht tp://www.tradeboss.com/default.cgi/action/viewcompan lobster, rock, spiny, demersal fish ies/searchterm/spiny+lobster/searchtermcondition/1/ , (snappers, groupers, grunts, ftp://ftp.fao.org/fi/DOCUMENT/IPOAS/national/Antigua U.S. LoF Caribbean spiny lobster trap/ pot >197 None documented, surgeonfish), flounder pots, traps 74 Lewis Fishing not applicable Antigua & Barbuda EEZ none documented none documented A/G AndBarbuda/NPOA_IUU.pdf Caribbean mixed species trap/pot are category III http://www.nmfs.noaa.gov/pr/interactions/fisheries/tabl lobster, rock, spiny free diving, loops 19 Lewis Fishing not applicable Antigua & Barbuda EEZ none documented none documented A/G e2/Atlantic_GOM_Caribbean_shellfish.html Queen conch (Strombus gigas), Dive (SCUBA & free molluscs diving) 25 not applicable not applicable Antigua & Barbuda EEZ none documented none documented A/G U.S. trade data Southeastern U.S. Atlantic, Gulf of Mexico, and Caribbean snapper- handline, hook and grouper and other reef fish bottom longline/hook-and-line/ >5,000 snapper line 71 Lewis Fishing not applicable Antigua & Barbuda EEZ none documented none documented N/I, A/G U.S. -

Effects of Acute and Chronic Air Exposure on Growth and Stress Response of Juvenile Olive Flounder, Paralichthys Olivaceus
www.trjfas.org ISSN 1303-2712 Turkish Journal of Fisheries and Aquatic Sciences 18:143-151 (2018) DOI: 10.4194/1303-2712-v18_1_16 RESEARCH PAPER Effects of Acute and Chronic Air Exposure on Growth and Stress Response of Juvenile Olive Flounder, Paralichthys olivaceus Han Kyu Lim1, Jun Wook Hur2,* 1Mokpo National University, Marine and Fisheries Resources, 1666 Youngsan-ro, Cheonggye, Muan, Jeonnam 58554, Korea. 2 Bio-Monitoring Center, 202 ho, 49, 1730 Beon-gil, Dongseodae-ro, Dong-gu, Daejeon, 300-805, Korea. * Corresponding Author: Tel.: +82.42 6386845; Fax: +82.42 6386845; Received 10 September 2016 E-mail: [email protected] Accepted 23 May 2017 Abstract We studied the effects of acute and chronic exposure to air on the growth and stress response of juvenile olive flounder, Paralichthys olivaceus. To study the stress response, the water was completely drained from the experimental tank, and the stressed group was exposed to air for 5 minutes, after which the tank was refilled with water. This stress was repeated daily for 30 days (between 1200 and 1300 h). From day 31 to day 69, no stress was applied. On day 70, the fish were again exposed to the air. The non-stressed group was not subjected to air exposure during the 70 days. We measured cortisol, glucose and lactic acid levels, osmolality, growth, survival, and feeding responses during the 70-day test period. Our results showed that olive flounder exhibit “typical” physiological responses (in cortisol, glucose, and lactic acid levels and osmolality) to the acute stress induced by air exposure. The response to chronic stress showed a similar increasing tendency. -

Recycled Fish Sculpture (.PDF)
Recycled Fish Sculpture Name:__________ Fish: are a paraphyletic group of organisms that consist of all gill-bearing aquatic vertebrate animals that lack limbs with digits. At 32,000 species, fish exhibit greater species diversity than any other group of vertebrates. Sculpture: is three-dimensional artwork created by shaping or combining hard materials—typically stone such as marble—or metal, glass, or wood. Softer ("plastic") materials can also be used, such as clay, textiles, plastics, polymers and softer metals. They may be assembled such as by welding or gluing or by firing, molded or cast. Researched Photo Source: Alaskan Rainbow STEP ONE: CHOOSE one fish from the attached Fish Names list. Trout STEP TWO: RESEARCH on-line and complete the attached K/U Fish Research Sheet. STEP THREE: DRAW 3 conceptual sketches with colour pencil crayons of possible visual images that represent your researched fish. STEP FOUR: Once your fish designs are approved by the teacher, DRAW a representational outline of your fish on the 18 x24 and then add VALUE and COLOUR . CONSIDER: Individual shapes and forms for the various parts you will cut out of recycled pop aluminum cans (such as individual scales, gills, fins etc.) STEP FIVE: CUT OUT using scissors the various individual sections of your chosen fish from recycled pop aluminum cans. OVERLAY them on top of your 18 x 24 Representational Outline 18 x 24 Drawing representational drawing to judge the shape and size of each piece. STEP SIX: Once you have cut out all your shapes and forms, GLUE the various pieces together with a glue gun. -

Betanodavirus and VER Disease: a 30-Year Research Review
pathogens Review Betanodavirus and VER Disease: A 30-year Research Review Isabel Bandín * and Sandra Souto Departamento de Microbioloxía e Parasitoloxía-Instituto de Acuicultura, Universidade de Santiago de Compostela, 15782 Santiago de Compostela, Spain; [email protected] * Correspondence: [email protected] Received: 20 December 2019; Accepted: 4 February 2020; Published: 9 February 2020 Abstract: The outbreaks of viral encephalopathy and retinopathy (VER), caused by nervous necrosis virus (NNV), represent one of the main infectious threats for marine aquaculture worldwide. Since the first description of the disease at the end of the 1980s, a considerable amount of research has gone into understanding the mechanisms involved in fish infection, developing reliable diagnostic methods, and control measures, and several comprehensive reviews have been published to date. This review focuses on host–virus interaction and epidemiological aspects, comprising viral distribution and transmission as well as the continuously increasing host range (177 susceptible marine species and epizootic outbreaks reported in 62 of them), with special emphasis on genotypes and the effect of global warming on NNV infection, but also including the latest findings in the NNV life cycle and virulence as well as diagnostic methods and VER disease control. Keywords: nervous necrosis virus (NNV); viral encephalopathy and retinopathy (VER); virus–host interaction; epizootiology; diagnostics; control 1. Introduction Nervous necrosis virus (NNV) is the causative agent of viral encephalopathy and retinopathy (VER), otherwise known as viral nervous necrosis (VNN). The disease was first described at the end of the 1980s in Australia and in the Caribbean [1–3], and has since caused a great deal of mortalities and serious economic losses in a variety of reared marine fish species, but also in freshwater species worldwide.