Peptide KRP Conjugated with Doxorubicin Exerts Anti-Tumor Activity by Regulating RPS6KA2 in Osteosarcoma
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Analysis of Gene Expression Data for Gene Ontology
ANALYSIS OF GENE EXPRESSION DATA FOR GENE ONTOLOGY BASED PROTEIN FUNCTION PREDICTION A Thesis Presented to The Graduate Faculty of The University of Akron In Partial Fulfillment of the Requirements for the Degree Master of Science Robert Daniel Macholan May 2011 ANALYSIS OF GENE EXPRESSION DATA FOR GENE ONTOLOGY BASED PROTEIN FUNCTION PREDICTION Robert Daniel Macholan Thesis Approved: Accepted: _______________________________ _______________________________ Advisor Department Chair Dr. Zhong-Hui Duan Dr. Chien-Chung Chan _______________________________ _______________________________ Committee Member Dean of the College Dr. Chien-Chung Chan Dr. Chand K. Midha _______________________________ _______________________________ Committee Member Dean of the Graduate School Dr. Yingcai Xiao Dr. George R. Newkome _______________________________ Date ii ABSTRACT A tremendous increase in genomic data has encouraged biologists to turn to bioinformatics in order to assist in its interpretation and processing. One of the present challenges that need to be overcome in order to understand this data more completely is the development of a reliable method to accurately predict the function of a protein from its genomic information. This study focuses on developing an effective algorithm for protein function prediction. The algorithm is based on proteins that have similar expression patterns. The similarity of the expression data is determined using a novel measure, the slope matrix. The slope matrix introduces a normalized method for the comparison of expression levels throughout a proteome. The algorithm is tested using real microarray gene expression data. Their functions are characterized using gene ontology annotations. The results of the case study indicate the protein function prediction algorithm developed is comparable to the prediction algorithms that are based on the annotations of homologous proteins. -

(PWP2) Gene Mapping to 21Q22.3 Ronald G
Downloaded from genome.cshlp.org on October 2, 2021 - Published by Cold Spring Harbor Laboratory Press LETTER Isolation and Genomic Structure of a Human Homolog of the Yeast Periodic Tryptophan Protein 2 (PWP2) Gene Mapping to 21q22.3 Ronald G. Lafrenii~re, 1'4 Daniel L. Rochefort, 1 Nathalie Chr~tien, ~ Catherine E. Neville, 2 Robert G. Korneluk, 2 Lin Zuo, 3 Yalin Wei, 3 Jay Lichter, 3 and Guy A. Rouleau ~ 1Centre for Research in Neuroscience, McGill University, and Department of Neurology, Montreal General Hospital Research Institute, Montreal, Canada H3G 1A4; 2DNA Sequencing Core Facility, Canadian Genetic Diseases Network, Children's Hospital of Eastern Ontario, Ottawa, Canada K1H 8L1; 3Sequana Therapeutics, Inc., La Jolla, California 92037 As part of efforts to identify candidate genes for diseases mapping to the 21q22.3 region, we have assembled a 770-kb cosmid and BAC contig containing eight tightly linked markers. These cosmids and BACs were restriction mapped using eight rare cutting enzymes, with the goal of identifying CpG-rich islands. One such island was identified by the clustering of lqotl, Eagl, Sstll, and BssHIl sites, and corresponded to the Nod linking clone LJI04 described previously. A 7.6-kb l-lindlll fragment containing this CpG-rich island was subcloned and partially sequenced. A homology search using the sequence obtained from either side of the Nod site identified an expressed sequence tag with homology to the yeast periodic tryptophan protein 2 (PWP2). Several cDNAs corresponding to the human PWP2 gene were identified and partially sequenced. Northern blot analysis revealed a 3.3-kb transcript that was well expressed in all tissues tested. -

Assembly and Annotation of an Ashkenazi Human Reference Genome
bioRxiv preprint doi: https://doi.org/10.1101/2020.03.18.997395; this version posted March 18, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Assembly and Annotation of an Ashkenazi Human Reference Genome Alaina Shumate1,2,† Aleksey V. Zimin1,2,† Rachel M. Sherman1,3 Daniela Puiu1,3 Justin M. Wagner4 Nathan D. Olson4 Mihaela Pertea1,2 Marc L. Salit5 Justin M. Zook4 Steven L. Salzberg1,2,3,6* 1Center for Computational Biology, Johns Hopkins University, Baltimore, MD 2Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD 3Department of Computer Science, Johns Hopkins University, Baltimore, MD 4National Institute of Standards and Technology, Gaithersburg, MD 5Joint Initiative for Metrology in Biology, Stanford University, Stanford, CA 6Department of Biostatistics, Johns Hopkins University, Baltimore, MD †These authors contributed equally to this work. *Corresponding author. Email: [email protected] Abstract Here we describe the assembly and annotation of the genome of an Ashkenazi individual and the creation of a new, population-specific human reference genome. This genome is more contiguous and more complete than GRCh38, the latest version of the human reference genome, and is annotated with highly similar gene content. The Ashkenazi reference genome, Ash1, contains 2,973,118,650 nucleotides as compared to 2,937,639,212 in GRCh38. Annotation identified 20,157 protein-coding genes, of which 19,563 are >99% identical to their counterparts on GRCh38. Most of the remaining genes have small differences. -

Supplementary Information Material and Methods
MCT-11-0474 BKM120: a potent and specific pan-PI3K inhibitor Supplementary Information Material and methods Chemicals The EGFR inhibitor NVP-AEE788 (Novartis), the Jak inhibitor I (Merck Calbiochem, #420099) and anisomycin (Alomone labs, # A-520) were prepared as 50 mM stock solutions in 100% DMSO. Doxorubicin (Adriablastin, Pfizer), EGF (Sigma Ref: E9644), PDGF (Sigma, Ref: P4306) and IL-4 (Sigma, Ref: I-4269) stock solutions were prepared as recommended by the manufacturer. For in vivo administration: Temodal (20 mg Temozolomide capsules, Essex Chemie AG, Luzern) was dissolved in 4 mL KZI/glucose (20/80, vol/vol); Taxotere was bought as 40 mg/mL solution (Sanofi Aventis, France), and prepared in KZI/glucose. Antibodies The primary antibodies used were as follows: anti-S473P-Akt (#9271), anti-T308P-Akt (#9276,), anti-S9P-GSK3β (#9336), anti-T389P-p70S6K (#9205), anti-YP/TP-Erk1/2 (#9101), anti-YP/TP-p38 (#9215), anti-YP/TP-JNK1/2 (#9101), anti-Y751P-PDGFR (#3161), anti- p21Cip1/Waf1 (#2946), anti-p27Kip1 (#2552) and anti-Ser15-p53 (#9284) antibodies were from Cell Signaling Technologies; anti-Akt (#05-591), anti-T32P-FKHRL1 (#06-952) and anti- PDGFR (#06-495) antibodies were from Upstate; anti-IGF-1R (#SC-713) and anti-EGFR (#SC-03) antibodies were from Santa Cruz; anti-GSK3α/β (#44610), anti-Y641P-Stat6 (#611566), anti-S1981P-ATM (#200-301), anti-T2609 DNA-PKcs (#GTX24194) and anti- 1 MCT-11-0474 BKM120: a potent and specific pan-PI3K inhibitor Y1316P-IGF-1R were from Bio-Source International, Becton-Dickinson, Rockland, GenTex and internal production, respectively. The 4G10 antibody was from Millipore (#05-321MG). -
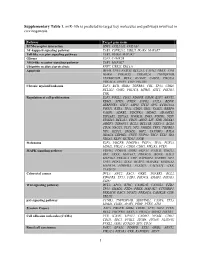
1 Supplementary Table 1. Mir-10B Is Predicted to Target Key Molecules
Supplementary Table 1. miR-10b is predicted to target key molecules and pathways involved in carcinogenesis. Pathway Target gene name ECM-receptor interaction SDC1, COL24A1, COL4A4 NF-kappa B signaling pathway TAB1, CSNK2A2, UBE2I, IRAK4, MAP3K7 Toll-like receptor signaling pathway TAB1, IRAK4, MAP3K7 Glioma E2F3, CAMK2B NOD-like receptor signaling pathway TAB1, MAP3K7 Ubiquitin mediated proteolysis RNF7, UBE2I, ERCC8 Apoptosis DFFB, TP53, FASLG, BCL2L1, CAPN2, PRKX, ATM, IRAK4, PRKACG, PRKAR2A, TNFRSF10B, TNFRSF10D, BCL2, IL1RAP, CASP8, PIK3CA, PRKACA, APAF1, CHP, PIK3R3 Chronic myeloid leukemia E2F3, BCR, GRB2, TGFBR1, CBL, TP53, CDK6, BCL2L1, GAB2, PIK3CA, MDM2, SHC1, PIK3R3, CRK Regulation of cell proliferation E2F3, FOSL2, CDX2, PDGFB, OSMR, E2F7, ARNT2, RBM5, STRN, PTEN, S1PR2, CUL3, BDNF, SERPINE1, SHC1, ASPH, ITCH, SPN, CCDC88A, FOXJ1, RXRA, TP53, CDK6, IRS1, VASH2, RBBP9, VASH1, ADRB2, PDGFRA, MDM2, ADAMTS1, EIF2AK2, EIF5A2, ICOSLG, ING5, FGFR3, NDN, ST8SIA1, BCL2L1, CDH5, ARNT, LIF, VDR, HOXA3, AGGF1, TSPAN31, BCL2, BCL11B, NKX3-1, BCL6, CD28, NACC1, FLT1, NF2, JARID2, TBX5, TGFBR1, NF1, KLF11, SMAD2, IGF2, TAX1BP3, BTLA, HDAC4, LEPRE1, CNTF, NUP62, TSC1, ETS1, ID4, NR5A2, KLF4, KCTD11, NFIB Melanoma E2F3, PDGFB, PDGFRA, FGF11, TP53, FGF23, MDM2, PIK3CA, CDK6, CDH1, PIK3R3, PTEN MAPK signaling pathway FGFR3, PDGFB, GRB2, FGF11, FASLG, GNG12, SRF, PRKX, MAP3K7, PRKACG, BDNF, RAC3, MAP3K2, PRKACA, CHP, RAPGEF2, TGFBR1, NF1, TP53, FGF23, STK4, DUSP5, MAP4K4, RPS6KA2, MAPK14, PDGFRA, PLA2G3, CACNA1C, CRK, PLA2G2F Colorectal cancer -

Small Nucleolar Rnas Determine Resistance to Doxorubicin in Human Osteosarcoma
International Journal of Molecular Sciences Article Small Nucleolar RNAs Determine Resistance to Doxorubicin in Human Osteosarcoma Martina Godel 1, Deborah Morena 1, Preeta Ananthanarayanan 1, Ilaria Buondonno 1, Giulio Ferrero 2,3 , Claudia M. Hattinger 4, Federica Di Nicolantonio 1,5 , Massimo Serra 4 , 1 2 1, , 1, , Riccardo Taulli , Francesca Cordero , Chiara Riganti * y and Joanna Kopecka * y 1 Department of Oncology, University of Torino, 1026 Torino, Italy; [email protected] (M.G.); [email protected] (D.M.); [email protected] (P.A.); [email protected] (I.B.); [email protected] (F.D.N.); [email protected] (R.T.) 2 Department of Computer Science, University of Torino, 10149 Torino, Italy; [email protected] (G.F.); [email protected] (F.C.) 3 Department of Clinical and Biological Sciences, University of Torino, 10043 Orbassano, Italy 4 Laboratory of Experimental Oncology, Pharmacogenomics and Pharmacogenetics Research Unit, IRCCS Istituto Ortopedico Rizzoli, 40136 Bologna, Italy; [email protected] (C.M.H.); [email protected] (M.S.) 5 Candiolo Cancer Institute, FPO–IRCCS, 10060 Candiolo, Italy * Correspondence: [email protected] (C.R.); [email protected] (J.K.); Tel.: +39-0116705857 (C.R.); +39-0116705849 (J.K.) These authors equally contributed to this work. y Received: 31 May 2020; Accepted: 21 June 2020; Published: 24 June 2020 Abstract: Doxorubicin (Dox) is one of the most important first-line drugs used in osteosarcoma therapy. Multiple and not fully clarified mechanisms, however, determine resistance to Dox. With the aim of identifying new markers associated with Dox-resistance, we found a global up-regulation of small nucleolar RNAs (snoRNAs) in human Dox-resistant osteosarcoma cells. -

Investigation of the Underlying Hub Genes and Molexular Pathogensis in Gastric Cancer by Integrated Bioinformatic Analyses
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.20.423656; this version posted December 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Investigation of the underlying hub genes and molexular pathogensis in gastric cancer by integrated bioinformatic analyses Basavaraj Vastrad1, Chanabasayya Vastrad*2 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2020.12.20.423656; this version posted December 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract The high mortality rate of gastric cancer (GC) is in part due to the absence of initial disclosure of its biomarkers. The recognition of important genes associated in GC is therefore recommended to advance clinical prognosis, diagnosis and and treatment outcomes. The current investigation used the microarray dataset GSE113255 RNA seq data from the Gene Expression Omnibus database to diagnose differentially expressed genes (DEGs). Pathway and gene ontology enrichment analyses were performed, and a proteinprotein interaction network, modules, target genes - miRNA regulatory network and target genes - TF regulatory network were constructed and analyzed. Finally, validation of hub genes was performed. The 1008 DEGs identified consisted of 505 up regulated genes and 503 down regulated genes. -

WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT (51) International Patent Classification: CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, C12Q 1/68 (2018.01) A61P 31/18 (2006.01) DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, C12Q 1/70 (2006.01) HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, (21) International Application Number: MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, PCT/US2018/056167 OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, (22) International Filing Date: SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, 16 October 2018 (16. 10.2018) TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (26) Publication Language: English GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, TZ, (30) Priority Data: UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, 62/573,025 16 October 2017 (16. 10.2017) US TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, EE, ES, FI, FR, GB, GR, HR, HU, ΓΕ , IS, IT, LT, LU, LV, (71) Applicant: MASSACHUSETTS INSTITUTE OF MC, MK, MT, NL, NO, PL, PT, RO, RS, SE, SI, SK, SM, TECHNOLOGY [US/US]; 77 Massachusetts Avenue, TR), OAPI (BF, BJ, CF, CG, CI, CM, GA, GN, GQ, GW, Cambridge, Massachusetts 02139 (US). -

Molecular Effects of Isoflavone Supplementation Human Intervention Studies and Quantitative Models for Risk Assessment
Molecular effects of isoflavone supplementation Human intervention studies and quantitative models for risk assessment Vera van der Velpen Thesis committee Promotors Prof. Dr Pieter van ‘t Veer Professor of Nutritional Epidemiology Wageningen University Prof. Dr Evert G. Schouten Emeritus Professor of Epidemiology and Prevention Wageningen University Co-promotors Dr Anouk Geelen Assistant professor, Division of Human Nutrition Wageningen University Dr Lydia A. Afman Assistant professor, Division of Human Nutrition Wageningen University Other members Prof. Dr Jaap Keijer, Wageningen University Dr Hubert P.J.M. Noteborn, Netherlands Food en Consumer Product Safety Authority Prof. Dr Yvonne T. van der Schouw, UMC Utrecht Dr Wendy L. Hall, King’s College London This research was conducted under the auspices of the Graduate School VLAG (Advanced studies in Food Technology, Agrobiotechnology, Nutrition and Health Sciences). Molecular effects of isoflavone supplementation Human intervention studies and quantitative models for risk assessment Vera van der Velpen Thesis submitted in fulfilment of the requirements for the degree of doctor at Wageningen University by the authority of the Rector Magnificus Prof. Dr M.J. Kropff, in the presence of the Thesis Committee appointed by the Academic Board to be defended in public on Friday 20 June 2014 at 13.30 p.m. in the Aula. Vera van der Velpen Molecular effects of isoflavone supplementation: Human intervention studies and quantitative models for risk assessment 154 pages PhD thesis, Wageningen University, Wageningen, NL (2014) With references, with summaries in Dutch and English ISBN: 978-94-6173-952-0 ABSTRact Background: Risk assessment can potentially be improved by closely linked experiments in the disciplines of epidemiology and toxicology. -

De Novo Genome Assembly and Analysis of Non- Allelic Recombination in Pathogenic Yeast Candida Glabrata
DE NOVO GENOME ASSEMBLY AND ANALYSIS OF NON- ALLELIC RECOMBINATION IN PATHOGENIC YEAST CANDIDA GLABRATA by Zhuwei Xu A dissertation submitted to Johns Hopkins University in conformity with the requirements for the degree of Doctor of Philosophy Baltimore, Maryland November 2019 © 2019 Zhuwei Xu All Rights Reserved Abstract Candida glabrata is an opportunistic pathogen in humans, responsible for approximately 20% of disseminated candidiasis. C. glabrata’s ability to adhere to host tissue is mediated by GPI- anchored cell wall proteins (GPI-CWPs); the corresponding genes contain long tandem repeat regions and form large gene families. These tandem repeats cause mis-assemblies of GPI-CWP genes in C. glabrata genome. Subtelomeres of C. glabrata are particularly rich in GPI-CWP genes and share homology with each other. Consequently, the subtelomeres are mis-assembled in genome sequences assembled from short sequencing reads. In this thesis, we used the long single-molecule real time (SMRT) reads and performed de novo genome assembly of the C. glabrata genome to establish the correct structure of GPI-CWP genes and the subtelomeres. We assembled the genome of six C. glabrata strains: the type strain, CBS138; our lab strain, BG2; four serial clinical isolates, BG3993-96 to assess genome changes during infection. With high quality sequences in hand, we then assess recombinational exchange between GPI-CWP genes by non-allelic mitotic recombination. This question is difficult to address with normal aligners, and we developed a k-mer based method to identify recombination. Our assembly established the correct subtelomere structure of Candida glabrata and provides correct structure of the GPI-CWP gene families. -

The P90 RSK Family Members: Common Functions and Isoform Specificity
Published OnlineFirst August 22, 2013; DOI: 10.1158/0008-5472.CAN-12-4448 Cancer Review Research The p90 RSK Family Members: Common Functions and Isoform Specificity Romain Lara, Michael J. Seckl, and Olivier E. Pardo Abstract The p90 ribosomal S6 kinases (RSK) are implicated in various cellular processes, including cell proliferation, survival, migration, and invasion. In cancer, RSKs modulate cell transformation, tumorigenesis, and metastasis. Indeed, changes in the expression of RSK isoforms have been reported in several malignancies, including breast, prostate, and lung cancers. Four RSK isoforms have been identified in humans on the basis of their high degree of sequence homology. Although this similarity suggests some functional redundancy between these proteins, an increasing body of evidence supports the existence of isoform-based specificity among RSKs in mediating particular cellular processes. This review briefly presents the similarities between RSK family members before focusing on the specific function of each of the isoforms and their involvement in cancer progression. Cancer Res; 73(17); 1–8. Ó2013 AACR. Introduction subsequently cloned throughout the Metazoan kingdom (2). The extracellular signal–regulated kinase (ERK)1/2 pathway The genomic analysis of several cancer types suggests that fi is involved in key cellular processes, including cell prolifera- these genes are not frequently ampli ed or mutated, with some tion, differentiation, survival, metabolism, and migration. notable exceptions (e.g., in the case of hepatocellular carcino- More than 30% of all human cancers harbor mutations within ma; ref. 6). Table 1 summarizes reported genetic changes in this pathway, mostly resulting in gain of function and conse- RSK genes. -

The Hominoid-Specific Gene TBC1D3 Promotes Generation of Basal Neural
1 The hominoid-specific gene TBC1D3 promotes generation of basal neural 2 progenitors and induces cortical folding in mice 3 4 Xiang-Chun Ju1,3,9, Qiong-Qiong Hou1,3,9, Ai-Li Sheng1, Kong-Yan Wu1, Yang Zhou8, 5 Ying Jin8, Tieqiao Wen7, Zhengang Yang6, Xiaoqun Wang2,5, Zhen-Ge Luo1,2,3,4 6 7 1Institute of Neuroscience, State Key Laboratory of Neuroscience, Shanghai Institutes for 8 Biological Sciences, Chinese Academy of Sciences, Shanghai, China. 9 2CAS Center for Excellence in Brain Science and Intelligence Technology, Shanghai, 10 China. 11 3Chinese Academy of Sciences University, Beijing, China. 12 4ShanghaiTech University, Shanghai, China 13 5Institute of Biophysics, Chinese Academy of Sciences, Beijing, China. 14 6Institutes of Brain Science, State Key Laboratory of Medical Neurobiology, Fudan 15 University, Shanghai, China. 16 7School of Life Sciences, Shanghai University, Shanghai, China. 17 8The Institute of Health Sciences, Shanghai Institutes for Biological Sciences, Chinese 18 Academy of Sciences, Shanghai, China. 19 9Co-first author 20 Correspondence should be addressed to Z.G.L ([email protected]) 21 22 Author contributions 23 X.-C.J and Q.-Q. H performed most experiments, analyzed data and wrote the paper. 24 A.-L.S helped with in situ hybridization. K.-Y.W assisted with imaging analysis. T.W 25 helped with the construction of nestin promoter construct. Y. Z and Y. J helped with 26 ReNeuron cell culture and analysis. Z.Y provided human fetal samples and assisted with 1 27 immunohistochemistry analysis. X.W provided help with live-imaging analysis. Z.-G.L 28 supervised the whole study, designed the research, analyzed data and wrote the paper.