Supplementary Information Material and Methods
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Evolution, Expression and Meiotic Behavior of Genes Involved in Chromosome Segregation of Monotremes
G C A T T A C G G C A T genes Article Evolution, Expression and Meiotic Behavior of Genes Involved in Chromosome Segregation of Monotremes Filip Pajpach , Linda Shearwin-Whyatt and Frank Grützner * School of Biological Sciences, The University of Adelaide, Adelaide, SA 5005, Australia; fi[email protected] (F.P.); [email protected] (L.S.-W.) * Correspondence: [email protected] Abstract: Chromosome segregation at mitosis and meiosis is a highly dynamic and tightly regulated process that involves a large number of components. Due to the fundamental nature of chromosome segregation, many genes involved in this process are evolutionarily highly conserved, but duplica- tions and functional diversification has occurred in various lineages. In order to better understand the evolution of genes involved in chromosome segregation in mammals, we analyzed some of the key components in the basal mammalian lineage of egg-laying mammals. The chromosome passenger complex is a multiprotein complex central to chromosome segregation during both mitosis and meio- sis. It consists of survivin, borealin, inner centromere protein, and Aurora kinase B or C. We confirm the absence of Aurora kinase C in marsupials and show its absence in both platypus and echidna, which supports the current model of the evolution of Aurora kinases. High expression of AURKBC, an ancestor of AURKB and AURKC present in monotremes, suggests that this gene is performing all necessary meiotic functions in monotremes. Other genes of the chromosome passenger complex complex are present and conserved in monotremes, suggesting that their function has been preserved Citation: Pajpach, F.; in mammals. -
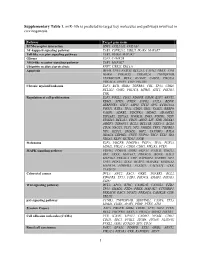
1 Supplementary Table 1. Mir-10B Is Predicted to Target Key Molecules
Supplementary Table 1. miR-10b is predicted to target key molecules and pathways involved in carcinogenesis. Pathway Target gene name ECM-receptor interaction SDC1, COL24A1, COL4A4 NF-kappa B signaling pathway TAB1, CSNK2A2, UBE2I, IRAK4, MAP3K7 Toll-like receptor signaling pathway TAB1, IRAK4, MAP3K7 Glioma E2F3, CAMK2B NOD-like receptor signaling pathway TAB1, MAP3K7 Ubiquitin mediated proteolysis RNF7, UBE2I, ERCC8 Apoptosis DFFB, TP53, FASLG, BCL2L1, CAPN2, PRKX, ATM, IRAK4, PRKACG, PRKAR2A, TNFRSF10B, TNFRSF10D, BCL2, IL1RAP, CASP8, PIK3CA, PRKACA, APAF1, CHP, PIK3R3 Chronic myeloid leukemia E2F3, BCR, GRB2, TGFBR1, CBL, TP53, CDK6, BCL2L1, GAB2, PIK3CA, MDM2, SHC1, PIK3R3, CRK Regulation of cell proliferation E2F3, FOSL2, CDX2, PDGFB, OSMR, E2F7, ARNT2, RBM5, STRN, PTEN, S1PR2, CUL3, BDNF, SERPINE1, SHC1, ASPH, ITCH, SPN, CCDC88A, FOXJ1, RXRA, TP53, CDK6, IRS1, VASH2, RBBP9, VASH1, ADRB2, PDGFRA, MDM2, ADAMTS1, EIF2AK2, EIF5A2, ICOSLG, ING5, FGFR3, NDN, ST8SIA1, BCL2L1, CDH5, ARNT, LIF, VDR, HOXA3, AGGF1, TSPAN31, BCL2, BCL11B, NKX3-1, BCL6, CD28, NACC1, FLT1, NF2, JARID2, TBX5, TGFBR1, NF1, KLF11, SMAD2, IGF2, TAX1BP3, BTLA, HDAC4, LEPRE1, CNTF, NUP62, TSC1, ETS1, ID4, NR5A2, KLF4, KCTD11, NFIB Melanoma E2F3, PDGFB, PDGFRA, FGF11, TP53, FGF23, MDM2, PIK3CA, CDK6, CDH1, PIK3R3, PTEN MAPK signaling pathway FGFR3, PDGFB, GRB2, FGF11, FASLG, GNG12, SRF, PRKX, MAP3K7, PRKACG, BDNF, RAC3, MAP3K2, PRKACA, CHP, RAPGEF2, TGFBR1, NF1, TP53, FGF23, STK4, DUSP5, MAP4K4, RPS6KA2, MAPK14, PDGFRA, PLA2G3, CACNA1C, CRK, PLA2G2F Colorectal cancer -

Poster Authors: Martin Lee Miller, Søren Brunak, Lars Juhl Jensen, Michael B
A Sequence-Specificity Atlas of the Kinase World CK1 family NEK2 Negative Atypical S TGFbR2 S D F T EIF2AK2 YS SY A EE S F EDDETDG S DTTYGA 0 1 2 3 4 5 6 7 -7 -6 -5 -4 -3 -2 -1 L I M S TR FWMW VS W G LT Y H T 0 1 2 3 4 5 6 7 -7 -6 -5 -4 -3 -2 -1 E S R CK1gamma3 CK1gamma1 CK1gamma2 R K T TS DE CK1epsilon S CK1alpha2 NND QRN S KKS M CQ CK1alpha 0 1 2 3 4 5 6 7 O -6 -5 -4 -3 -2 -1 T CK1delta -7 V D KRT R D E RL D R SEL T PTTS LD T S S L S SK LM PLP LS E E P A E K RG LDLF Y G V N G GR L I A T LV VAY TT TTBK2 KR I T RVFS I RE KA TDD RS KQ KM D EE G G BUBR1 TTBK1 VRK2 VRK1 N RE P D SgK396 VRK3 MPKRT F NFNMGPM SgK196 P I GMVHG KYL I MEL SgK493 Positive 0 SgK110 Haspin S BUB1 -7 -6 -5 -4 -3 -2 -1 SgK069 +3 +4 +5 +6 +7 +1 +2 SgK223 SgK269 Wnk2 Wnk4 Wnk3 NRBP1 ATM/ATR NRBP2 Wnk1 SCYL2 SCYL1 SCYL3 PIK3R4 Slob SgK424 SgK307 PINK1 SBK SgK496 MOS TBCK PBK CYGF CDC7 CYGD HSER NEK1 ANPb Proline NEK5 ANPa ILK NEK3 BMPR2 NEK11NEK4 MISR2 TGFbR2 ACTR2 NEK2 ACTR2B ALK2 NEK9 ALK1 NEK8 BMPR1B NEK6 BMPR1A NEK10NEK7 ALK7 TGFbR1 ALK4 AAK1 MLKL IRAK4 S S MPSK1 BIKE IRAK2 E Q EIF2AK2 S D G S E IRAK1 ES A S PS I SS GAK TQL PL SQ DL Syk family S VP Tec family P D E IRAK3 EphB3 L T GCN2 1 TESK2 0 PEK TESK1 -7 -6 -5 -4 -3 -2 -1 S +1 +2 +3 +4 +5 +6 +7 Q LIMK1 Wee1B LIMK2 mTOR LRRK2 HRI LRRK1 Wee1 RIPK1 MYT1 RIPK3 RNAseL RIPK2 IRE1 ANKRD3 IRE2 SgK288 KSR2 SgK071 KSR1 D E V L P KIS ARAF EV P S D R TTK E D D R VE AD VS A EE D DG V SSDDE EY NP R BRAF D V L E M ATPVD CLIK1LCLIK1 S EES E YE L LEE QLT EN PD E A L D T I SP NQ DG ANSS D DEE S DLL MS G PS P K A SD TEN T NGLKND -

Profiling Data
Compound Name DiscoveRx Gene Symbol Entrez Gene Percent Compound Symbol Control Concentration (nM) JNK-IN-8 AAK1 AAK1 69 1000 JNK-IN-8 ABL1(E255K)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317I)-nonphosphorylated ABL1 87 1000 JNK-IN-8 ABL1(F317I)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317L)-nonphosphorylated ABL1 65 1000 JNK-IN-8 ABL1(F317L)-phosphorylated ABL1 61 1000 JNK-IN-8 ABL1(H396P)-nonphosphorylated ABL1 42 1000 JNK-IN-8 ABL1(H396P)-phosphorylated ABL1 60 1000 JNK-IN-8 ABL1(M351T)-phosphorylated ABL1 81 1000 JNK-IN-8 ABL1(Q252H)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(Q252H)-phosphorylated ABL1 56 1000 JNK-IN-8 ABL1(T315I)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(T315I)-phosphorylated ABL1 92 1000 JNK-IN-8 ABL1(Y253F)-phosphorylated ABL1 71 1000 JNK-IN-8 ABL1-nonphosphorylated ABL1 97 1000 JNK-IN-8 ABL1-phosphorylated ABL1 100 1000 JNK-IN-8 ABL2 ABL2 97 1000 JNK-IN-8 ACVR1 ACVR1 100 1000 JNK-IN-8 ACVR1B ACVR1B 88 1000 JNK-IN-8 ACVR2A ACVR2A 100 1000 JNK-IN-8 ACVR2B ACVR2B 100 1000 JNK-IN-8 ACVRL1 ACVRL1 96 1000 JNK-IN-8 ADCK3 CABC1 100 1000 JNK-IN-8 ADCK4 ADCK4 93 1000 JNK-IN-8 AKT1 AKT1 100 1000 JNK-IN-8 AKT2 AKT2 100 1000 JNK-IN-8 AKT3 AKT3 100 1000 JNK-IN-8 ALK ALK 85 1000 JNK-IN-8 AMPK-alpha1 PRKAA1 100 1000 JNK-IN-8 AMPK-alpha2 PRKAA2 84 1000 JNK-IN-8 ANKK1 ANKK1 75 1000 JNK-IN-8 ARK5 NUAK1 100 1000 JNK-IN-8 ASK1 MAP3K5 100 1000 JNK-IN-8 ASK2 MAP3K6 93 1000 JNK-IN-8 AURKA AURKA 100 1000 JNK-IN-8 AURKA AURKA 84 1000 JNK-IN-8 AURKB AURKB 83 1000 JNK-IN-8 AURKB AURKB 96 1000 JNK-IN-8 AURKC AURKC 95 1000 JNK-IN-8 -

Glycogen Synthase Kinase-3 Is Activated in Neuronal Cells by G 12
The Journal of Neuroscience, August 15, 2002, 22(16):6863–6875 Glycogen Synthase Kinase-3 Is Activated in Neuronal Cells by G␣ ␣ 12 and G 13 by Rho-Independent and Rho-Dependent Mechanisms C. Laura Sayas, Jesu´ s Avila, and Francisco Wandosell Centro de Biologı´a Molecular “Severo Ochoa”, Consejo Superior de Investigaciones Cientı´ficas, Universidad Auto´ noma de Madrid, Cantoblanco, Madrid 28049, Spain ␣ ␣ ␣ ␣ Glycogen synthase kinase-3 (GSK-3) was generally considered tively active G 12 (G 12QL) and G 13 (G 13QL) in Neuro2a cells a constitutively active enzyme, only regulated by inhibition. induces upregulation of GSK-3 activity. Furthermore, overex- Here we describe that GSK-3 is activated by lysophosphatidic pression of constitutively active RhoA (RhoAV14) also activates ␣ acid (LPA) during neurite retraction in rat cerebellar granule GSK-3 However, the activation of GSK-3 by G 13 is blocked by neurons. GSK-3 activation correlates with an increase in GSK-3 coexpression with C3 transferase, whereas C3 does not block ␣ tyrosine phosphorylation. In addition, LPA induces a GSK-3- GSK-3 activation by G 12. Thus, we demonstrate that GSK-3 is ␣ ␣ mediated hyperphosphorylation of the microtubule-associated activated by both G 12 and G 13 in neuronal cells. However, ␣ protein tau. Inhibition of GSK-3 by lithium partially blocks neu- GSK-3 activation by G 13 is Rho-mediated, whereas GSK-3 ␣ rite retraction, indicating that GSK-3 activation is important but activation by G 12 is Rho-independent. The results presented not essential for the neurite retraction progress. GSK-3 activa- here imply the existence of a previously unknown mechanism of ␣ tion by LPA in cerebellar granule neurons is neither downstream GSK-3 activation by G 12/13 subunits. -

Multistep Regulation of Autophagy by WNK1
Multistep regulation of autophagy by WNK1 Sachith Gallolu Kankanamalagea, A-Young Leea, Chonlarat Wichaidita, Andres Lorente-Rodrigueza, Akansha M. Shaha, Steve Stippeca, Angelique W. Whitehurstb, and Melanie H. Cobba,b,1 aDepartment of Pharmacology, University of Texas Southwestern Medical Center, Dallas, TX 75390; and bHarold C. Simmons Comprehensive Cancer Center, University of Texas Southwestern Medical Center, Dallas, TX 75390 Contributed by Melanie H. Cobb, October 27, 2016 (sent for review September 6, 2016; reviewed by Jing Liu and Helen Piwnica-Worms) The with-no-lysine (K) (WNK) kinases are an atypical family of components and to supply cells with nutrients and building blocks protein kinases that regulate ion transport across cell membranes. (30–33). Autophagy is induced by cellular stress and protects Mutations that result in their overexpression cause hypertension- against infections by pathogens (34–40). Critical to maintain in- related disorders in humans. Of the four mammalian WNKs, only tracellular homeostasis, autophagy has roles in diseases, such as WNK1 is expressed throughout the body. We report that WNK1 neurodegeneration (41, 42) and cancer (43, 44). In this study, we inhibits autophagy, an intracellular degradation pathway impli- show that WNK1 is involved in regulating autophagy. cated in several human diseases. Using small-interfering RNA- mediated WNK1 knockdown, we show autophagosome formation Results and autophagic flux are accelerated. In cells with reduced WNK1, WNK1 Depletion Increases Autophagy. To analyze its role in auto- basal and starvation-induced autophagy is increased. We also phagy, WNK1 was knocked down with small interfering RNA show that depletion of WNK1 stimulates focal class III phospha- (siRNA) in U2OS cells stably expressing green fluorescent protein- tidylinositol 3-kinase complex (PI3KC3) activity, which is required tagged light chain 3 (GFP-LC3) (32, 45). -

Application of a MYC Degradation
SCIENCE SIGNALING | RESEARCH ARTICLE CANCER Copyright © 2019 The Authors, some rights reserved; Application of a MYC degradation screen identifies exclusive licensee American Association sensitivity to CDK9 inhibitors in KRAS-mutant for the Advancement of Science. No claim pancreatic cancer to original U.S. Devon R. Blake1, Angelina V. Vaseva2, Richard G. Hodge2, McKenzie P. Kline3, Thomas S. K. Gilbert1,4, Government Works Vikas Tyagi5, Daowei Huang5, Gabrielle C. Whiten5, Jacob E. Larson5, Xiaodong Wang2,5, Kenneth H. Pearce5, Laura E. Herring1,4, Lee M. Graves1,2,4, Stephen V. Frye2,5, Michael J. Emanuele1,2, Adrienne D. Cox1,2,6, Channing J. Der1,2* Stabilization of the MYC oncoprotein by KRAS signaling critically promotes the growth of pancreatic ductal adeno- carcinoma (PDAC). Thus, understanding how MYC protein stability is regulated may lead to effective therapies. Here, we used a previously developed, flow cytometry–based assay that screened a library of >800 protein kinase inhibitors and identified compounds that promoted either the stability or degradation of MYC in a KRAS-mutant PDAC cell line. We validated compounds that stabilized or destabilized MYC and then focused on one compound, Downloaded from UNC10112785, that induced the substantial loss of MYC protein in both two-dimensional (2D) and 3D cell cultures. We determined that this compound is a potent CDK9 inhibitor with a previously uncharacterized scaffold, caused MYC loss through both transcriptional and posttranslational mechanisms, and suppresses PDAC anchorage- dependent and anchorage-independent growth. We discovered that CDK9 enhanced MYC protein stability 62 through a previously unknown, KRAS-independent mechanism involving direct phosphorylation of MYC at Ser . -

The Number of Genes
Table S1. The numbers of KD genes in each KD time The number The number The number The number Cell lines of genes of genes of genes of genes (96h) (120h) (144h) PC3 3980 3822 128 1725 A549 3724 3724 0 0 MCF7 3688 3471 0 1837 HT29 3665 3665 0 0 A375 3826 3826 0 0 HA1E 3801 3801 0 0 VCAP 4134 34 4121 0 HCC515 3522 3522 0 0 Table S2. The predicted results in the PC3 cell line on the LINCS II data id target rank A07563059 ADRB2 48 A12896037 ADRA2C 91 A13021932 YES1 77 PPM1B;PPP1CC;PPP2CA; A13254067 584;1326;297;171;3335 PTPN1;PPP2R5A A16347691 GMNN 2219 PIK3CB;MTOR;PIK3CA;PIK A28467416 18;10;9;13;8 3CG;PIK3CD A28545468 EHMT2;MAOB 14;67 A29520968 HSPB1 1770 A48881734 EZH2 1596 A52922642 CACNA1C 201 A64553394 ADRB2 155 A65730376 DOT1L 3764 A82035391 JUN 378 A82156122 DPP4 771 HRH1;HTR2C;CHRM3;CH A82772293 2756;2354;2808;2367 RM1 A86248581 CDA 1785 A92800748 TEK 459 A93093700 LMNA 1399 K00152668 RARB 105 K01577834 ADORA2A 525 K01674964 HRH1;BLM 31;1314 K02314383 AR 132 K03194791 PDE4D 30 K03390685 MAP2K1 77 K06762493 GMNN;APEX1 1523;2360 K07106112 ERBB4;ERBB2;EGFR 497;60;23 K07310275 AKT1;MTOR;PIK3CA 13;12;1 K07753030 RGS4;BLM 3736;3080 K08109215 BRD2;BRD3;BRD4 1413;2786;3 K08248804 XIAP 88 K08586861 TBXA2R;MBNL1 297;3428 K08832567 GMNN;CA12 2544;50 LMNA;NFKB1;APEX1;EH K08976401 1322;341;3206;123 MT2 K09372874 IMPDH2 232 K09711437 PLA2G2A 59 K10859802 GPR119 214 K11267252 RET;ALK 395;760 K12609457 LMNA 907 K13094524 BRD4 7 K13662825 CDK4;CDK9;CDK5;CDK1 34;58;13;18 K14704277 LMNA;BLM 1697;1238 K14870255 AXL 1696 K15170068 MAN2B1 1756 K15179879 -

Kinase Profiling Book
Custom and Pre-Selected Kinase Prof iling to f it your Budget and Needs! As of July 1, 2021 19.8653 mm 128 196 12 Tyrosine Serine/Threonine Lipid Kinases Kinases Kinases Carna Biosciences, Inc. 2007 Carna Biosciences, Inc. Profiling Assays available from Carna Biosciences, Inc. As of July 1, 2021 Page Kinase Name Assay Platform Page Kinase Name Assay Platform 4 ABL(ABL1) MSA 21 EGFR[T790M/C797S/L858R] MSA 4 ABL(ABL1)[E255K] MSA 21 EGFR[T790M/L858R] MSA 4 ABL(ABL1)[T315I] MSA 21 EPHA1 MSA 4 ACK(TNK2) MSA 21 EPHA2 MSA 4 AKT1 MSA 21 EPHA3 MSA 5 AKT2 MSA 22 EPHA4 MSA 5 AKT3 MSA 22 EPHA5 MSA 5 ALK MSA 22 EPHA6 MSA 5 ALK[C1156Y] MSA 22 EPHA7 MSA 5 ALK[F1174L] MSA 22 EPHA8 MSA 6 ALK[G1202R] MSA 23 EPHB1 MSA 6 ALK[G1269A] MSA 23 EPHB2 MSA 6 ALK[L1196M] MSA 23 EPHB3 MSA 6 ALK[R1275Q] MSA 23 EPHB4 MSA 6 ALK[T1151_L1152insT] MSA 23 Erk1(MAPK3) MSA 7 EML4-ALK MSA 24 Erk2(MAPK1) MSA 7 NPM1-ALK MSA 24 Erk5(MAPK7) MSA 7 AMPKα1/β1/γ1(PRKAA1/B1/G1) MSA 24 FAK(PTK2) MSA 7 AMPKα2/β1/γ1(PRKAA2/B1/G1) MSA 24 FER MSA 7 ARG(ABL2) MSA 24 FES MSA 8 AurA(AURKA) MSA 25 FGFR1 MSA 8 AurA(AURKA)/TPX2 MSA 25 FGFR1[V561M] MSA 8 AurB(AURKB)/INCENP MSA 25 FGFR2 MSA 8 AurC(AURKC) MSA 25 FGFR2[V564I] MSA 8 AXL MSA 25 FGFR3 MSA 9 BLK MSA 26 FGFR3[K650E] MSA 9 BMX MSA 26 FGFR3[K650M] MSA 9 BRK(PTK6) MSA 26 FGFR3[V555L] MSA 9 BRSK1 MSA 26 FGFR3[V555M] MSA 9 BRSK2 MSA 26 FGFR4 MSA 10 BTK MSA 27 FGFR4[N535K] MSA 10 BTK[C481S] MSA 27 FGFR4[V550E] MSA 10 BUB1/BUB3 MSA 27 FGFR4[V550L] MSA 10 CaMK1α(CAMK1) MSA 27 FGR MSA 10 CaMK1δ(CAMK1D) MSA 27 FLT1 MSA 11 CaMK2α(CAMK2A) MSA 28 -

Lipid Metabolic Reprogramming: Role in Melanoma Progression and Therapeutic Perspectives
cancers Review Lipid metabolic Reprogramming: Role in Melanoma Progression and Therapeutic Perspectives 1, 1, 1 2 1 Laurence Pellerin y, Lorry Carrié y , Carine Dufau , Laurence Nieto , Bruno Ségui , 1,3 1, , 1, , Thierry Levade , Joëlle Riond * z and Nathalie Andrieu-Abadie * z 1 Centre de Recherches en Cancérologie de Toulouse, Equipe Labellisée Fondation ARC, Université Fédérale de Toulouse Midi-Pyrénées, Université Toulouse III Paul-Sabatier, Inserm 1037, 2 avenue Hubert Curien, tgrCS 53717, 31037 Toulouse CEDEX 1, France; [email protected] (L.P.); [email protected] (L.C.); [email protected] (C.D.); [email protected] (B.S.); [email protected] (T.L.) 2 Institut de Pharmacologie et de Biologie Structurale, CNRS, Université Toulouse III Paul-Sabatier, UMR 5089, 205 Route de Narbonne, 31400 Toulouse, France; [email protected] 3 Laboratoire de Biochimie Métabolique, CHU Toulouse, 31059 Toulouse, France * Correspondence: [email protected] (J.R.); [email protected] (N.A.-A.); Tel.: +33-582-7416-20 (J.R.) These authors contributed equally to this work. y These authors jointly supervised this work. z Received: 15 September 2020; Accepted: 23 October 2020; Published: 27 October 2020 Simple Summary: Melanoma is a devastating skin cancer characterized by an impressive metabolic plasticity. Melanoma cells are able to adapt to the tumor microenvironment by using a variety of fuels that contribute to tumor growth and progression. In this review, the authors summarize the contribution of the lipid metabolic network in melanoma plasticity and aggressiveness, with a particular attention to specific lipid classes such as glycerophospholipids, sphingolipids, sterols and eicosanoids. -

Article Reference
Article Phosphorylation by NLK inhibits YAP‐14‐3‐3‐interactions and induces its nuclear localization MOON, Sungho, et al. Reference MOON, Sungho, et al. Phosphorylation by NLK inhibits YAP‐14‐3‐3‐interactions and induces its nuclear localization. EMBO Reports, 2017, vol. 18, no. 1, p. 61-71 PMID : 27979972 DOI : 10.15252/embr.201642683 Available at: http://archive-ouverte.unige.ch/unige:112477 Disclaimer: layout of this document may differ from the published version. 1 / 1 Published online: December 15, 2016 Scientific Report Phosphorylation by NLK inhibits YAP-14-3-3- interactions and induces its nuclear localization Sungho Moon1,† , Wantae Kim1,†, Soyoung Kim1, Youngeun Kim1, Yonghee Song1, Oleksii Bilousov2, Jiyoung Kim1, Taebok Lee1, Boksik Cha1, Minseong Kim1, Hanjun Kim1, Vladimir L Katanaev2,3,* & Eek-hoon Jho1,** Abstract tissue homeostasis have become a long-standing topic of interest. As loss of the organ size control is linked to many human diseases, includ- Hippo signaling controls organ size by regulating cell proliferation ing cancer and degenerative diseases, regulation of the organ size and apoptosis. Yes-associated protein (YAP) is a key downstream could be an attractive therapeutic strategy. Recently, Hippo signaling effector of Hippo signaling, and LATS-mediated phosphorylation of has been identified as a major signaling pathway to control the organ YAP at Ser127 inhibits its nuclear localization and transcriptional size; dysregulation of this pathway results in aberrant growth [1]. activity. Here, we report that Nemo-like kinase (NLK) phosphory- The Hippo pathway is evolutionarily conserved from nematodes lates YAP at Ser128 both in vitro and in vivo, which blocks interac- to humans and controls a variety of cellular processes, such as cell tion with 14-3-3 and enhances its nuclear localization. -

Phosphorylation of Threonine 3 on Histone H3 by Haspin Kinase Is Required for Meiosis I in Mouse Oocytes
ß 2014. Published by The Company of Biologists Ltd | Journal of Cell Science (2014) 127, 5066–5078 doi:10.1242/jcs.158840 RESEARCH ARTICLE Phosphorylation of threonine 3 on histone H3 by haspin kinase is required for meiosis I in mouse oocytes Alexandra L. Nguyen, Amanda S. Gentilello, Ahmed Z. Balboula*, Vibha Shrivastava, Jacob Ohring and Karen Schindler` ABSTRACT a lesser extent. However, it is not known whether haspin is required for meiosis in oocytes. To date, the only known haspin substrates Meiosis I (MI), the division that generates haploids, is prone to are threonine 3 of histone H3 (H3T3), serine 137 of macroH2A and errors that lead to aneuploidy in females. Haspin is a kinase that threonine 57 of CENP-T (Maiolica et al., 2014). Knockdown or phosphorylates histone H3 on threonine 3, thereby recruiting Aurora inhibition of haspin in mitotically dividing tissue culture cell kinase B (AURKB) and the chromosomal passenger complex lines reveal that phosphorylation of H3T3 is essential for the (CPC) to kinetochores to regulate mitosis. Haspin and AURKC, an alignment of chromosomes at the metaphase plate (Dai and AURKB homolog, are enriched in germ cells, yet their significance Higgins, 2005; Dai et al., 2005), regulation of chromosome in regulating MI is not fully understood. Using inhibitors and cohesion (Dai et al., 2009) and establishing a bipolar spindle (Dai overexpression approaches, we show a role for haspin during MI et al., 2009). In mitotic metaphase, phosphorylation of H3T3 is in mouse oocytes. Haspin-perturbed oocytes display abnormalities restricted to kinetochores, and this mark signals recruitment of the in chromosome morphology and alignment, improper kinetochore– chromosomal passenger complex (CPC) (Dai et al., 2005; Wang microtubule attachments at metaphase I and aneuploidy at et al., 2010; Yamagishi et al., 2010).