Kinase Profiling Book
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplemental Information to Mammadova-Bach Et Al., “Laminin Α1 Orchestrates VEGFA Functions in the Ecosystem of Colorectal Carcinogenesis”
Supplemental information to Mammadova-Bach et al., “Laminin α1 orchestrates VEGFA functions in the ecosystem of colorectal carcinogenesis” Supplemental material and methods Cloning of the villin-LMα1 vector The plasmid pBS-villin-promoter containing the 3.5 Kb of the murine villin promoter, the first non coding exon, 5.5 kb of the first intron and 15 nucleotides of the second villin exon, was generated by S. Robine (Institut Curie, Paris, France). The EcoRI site in the multi cloning site was destroyed by fill in ligation with T4 polymerase according to the manufacturer`s instructions (New England Biolabs, Ozyme, Saint Quentin en Yvelines, France). Site directed mutagenesis (GeneEditor in vitro Site-Directed Mutagenesis system, Promega, Charbonnières-les-Bains, France) was then used to introduce a BsiWI site before the start codon of the villin coding sequence using the 5’ phosphorylated primer: 5’CCTTCTCCTCTAGGCTCGCGTACGATGACGTCGGACTTGCGG3’. A double strand annealed oligonucleotide, 5’GGCCGGACGCGTGAATTCGTCGACGC3’ and 5’GGCCGCGTCGACGAATTCACGC GTCC3’ containing restriction site for MluI, EcoRI and SalI were inserted in the NotI site (present in the multi cloning site), generating the plasmid pBS-villin-promoter-MES. The SV40 polyA region of the pEGFP plasmid (Clontech, Ozyme, Saint Quentin Yvelines, France) was amplified by PCR using primers 5’GGCGCCTCTAGATCATAATCAGCCATA3’ and 5’GGCGCCCTTAAGATACATTGATGAGTT3’ before subcloning into the pGEMTeasy vector (Promega, Charbonnières-les-Bains, France). After EcoRI digestion, the SV40 polyA fragment was purified with the NucleoSpin Extract II kit (Machery-Nagel, Hoerdt, France) and then subcloned into the EcoRI site of the plasmid pBS-villin-promoter-MES. Site directed mutagenesis was used to introduce a BsiWI site (5’ phosphorylated AGCGCAGGGAGCGGCGGCCGTACGATGCGCGGCAGCGGCACG3’) before the initiation codon and a MluI site (5’ phosphorylated 1 CCCGGGCCTGAGCCCTAAACGCGTGCCAGCCTCTGCCCTTGG3’) after the stop codon in the full length cDNA coding for the mouse LMα1 in the pCIS vector (kindly provided by P. -

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

Evolution, Expression and Meiotic Behavior of Genes Involved in Chromosome Segregation of Monotremes
G C A T T A C G G C A T genes Article Evolution, Expression and Meiotic Behavior of Genes Involved in Chromosome Segregation of Monotremes Filip Pajpach , Linda Shearwin-Whyatt and Frank Grützner * School of Biological Sciences, The University of Adelaide, Adelaide, SA 5005, Australia; fi[email protected] (F.P.); [email protected] (L.S.-W.) * Correspondence: [email protected] Abstract: Chromosome segregation at mitosis and meiosis is a highly dynamic and tightly regulated process that involves a large number of components. Due to the fundamental nature of chromosome segregation, many genes involved in this process are evolutionarily highly conserved, but duplica- tions and functional diversification has occurred in various lineages. In order to better understand the evolution of genes involved in chromosome segregation in mammals, we analyzed some of the key components in the basal mammalian lineage of egg-laying mammals. The chromosome passenger complex is a multiprotein complex central to chromosome segregation during both mitosis and meio- sis. It consists of survivin, borealin, inner centromere protein, and Aurora kinase B or C. We confirm the absence of Aurora kinase C in marsupials and show its absence in both platypus and echidna, which supports the current model of the evolution of Aurora kinases. High expression of AURKBC, an ancestor of AURKB and AURKC present in monotremes, suggests that this gene is performing all necessary meiotic functions in monotremes. Other genes of the chromosome passenger complex complex are present and conserved in monotremes, suggesting that their function has been preserved Citation: Pajpach, F.; in mammals. -

1 Tumor Suppressor PLK2 May Serve As a Biomarker in Triple-Negative Breast Cancer for Improved Response to PLK1 Therapeutics
bioRxiv preprint doi: https://doi.org/10.1101/2021.06.16.448722; this version posted June 16, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Tumor suppressor PLK2 may serve as a biomarker in triple-negative breast cancer for improved response to PLK1 therapeutics Yang Gao1, 2, 3, Elena B. Kabotyanski1, 2, Elizabeth Villegas7, Jonathan H. Shepherd8, Deanna Acosta1, 2, Clark Hamor1, 2, Tingting Sun2,4,5, Celina Montmeyor-Garcia9, Xiaping He8, Lacey E. Dobrolecki1, 2, 3, Thomas F. Westbrook2, 4, 5, Michael T. Lewis1, 2, 3, Susan G. Hilsenbeck2, 3, Xiang H.-F. Zhang1, 2, 3, 6, Charles M. Perou8 and Jeffrey M. Rosen1, 2 1Department of Molecular and Cellular Biology 2Dan L. Duncan Cancer Center 3Lester and Sue Smith Breast Center 4Department of Molecular and Human Genetics 5Verna & Marrs McLean Department of Biochemistry and Molecular Biology 6McNair Medical Institute Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030, USA 7University of Houston-Downtown, Houston, TX 77002, USA 8The University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA 9 Canadian Blood Services, Toronto, ON M5G 2M1, Canada Correspondence to Jeffrey M. Rosen (Mail Stop: BCM130, Room: BCM-M638a, Baylor College of Medicine, 1 Baylor Plaza, Houston, TX 77030. Office: 713-798-6210. Fax: 713-898-8012. Email: [email protected]) 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.06.16.448722; this version posted June 16, 2021. -

Epithelial Ovarian Cancer: Searching for New Modulators of Drug Resistance
UNIVERSITÀ DEGLI STUDI DI UDINE Dipartimento di scienze mediche e biologiche PhD COURSE IN BIOMEDICAL SCIENCES AND BIOTECHNOLOGY XXIX cycle PhD THESIS Epithelial Ovarian Cancer: searching for new modulators of drug resistance. PhD student Supervisor Prof. Carlo Ennio Michele Pucillo Valentina Ranzuglia Co-supervisors Dr. Gustavo Baldassarre Dr. Monica Schiappacassi PhD coordinator Prof. Claudio Brancolini Academic year 2015/2016 This PhD work was done at Centro di Riferimento Oncologico (CRO, National Cancer Institute ) of Aviano, in the Division of Experimental Oncology 2 directed by Dr. Gustavo Baldassarre. i Table of contents Abstract 3 1. Introduction 4 1.1 Epithelial Ovarian Cancer 4 1.1.1 Platinum resistance 5 1.2 Functional Genomic Screening 8 1.3 Serum and glucocorticoid-regulated kinases 9 1.3.1 SGK structure 10 1.3.2 SGK activation and regulation 11 1.3.3 Role of SGKs in regulation of molecular and cellular functions 13 1.3.4 Serum- and glucocorticoid-regulated kinases in cancer 15 1.4 Autophagy 17 2.Aim of the study 21 3.Results 22 3.1 Identification of SGK2 as a mediator of platinum sensitivity in EOC cells. 22 3.2 SGK2 silencing sensitizes EOC cells to platinum. 23 3.3 SGK2 overexpression confers an increased resistance to platinum drug. 25 3.4 SGK2-overexpressing cells present an increased in vitro and in vivo growth 26 rate 3.5 SGK2 kinase activity is involved in EOC sensitivity to platinum treatment. 28 3.6 The SGK1/SGK2 kinase inhibitor, GSK650394, reduces cell viability in 29 combination with platinum. 3.7 GSK650394 is able to make SGK2-expressing cells more sensitive to platinum. -

AGC Kinases in Mtor Signaling, in Mike Hall and Fuyuhiko Tamanoi: the Enzymes, Vol
Provided for non-commercial research and educational use only. Not for reproduction, distribution or commercial use. This chapter was originally published in the book, The Enzymes, Vol .27, published by Elsevier, and the attached copy is provided by Elsevier for the author's benefit and for the benefit of the author's institution, for non-commercial research and educational use including without limitation use in instruction at your institution, sending it to specific colleagues who know you, and providing a copy to your institution’s administrator. All other uses, reproduction and distribution, including without limitation commercial reprints, selling or licensing copies or access, or posting on open internet sites, your personal or institution’s website or repository, are prohibited. For exceptions, permission may be sought for such use through Elsevier's permissions site at: http://www.elsevier.com/locate/permissionusematerial From: ESTELA JACINTO, AGC Kinases in mTOR Signaling, In Mike Hall and Fuyuhiko Tamanoi: The Enzymes, Vol. 27, Burlington: Academic Press, 2010, pp.101-128. ISBN: 978-0-12-381539-2, © Copyright 2010 Elsevier Inc, Academic Press. Author's personal copy 7 AGC Kinases in mTOR Signaling ESTELA JACINTO Department of Physiology and Biophysics UMDNJ-Robert Wood Johnson Medical School, Piscataway New Jersey, USA I. Abstract The mammalian target of rapamycin (mTOR), a protein kinase with homology to lipid kinases, orchestrates cellular responses to growth and stress signals. Various extracellular and intracellular inputs to mTOR are known. mTOR processes these inputs as part of two mTOR protein com- plexes, mTORC1 or mTORC2. Surprisingly, despite the many cellular functions that are linked to mTOR, there are very few direct mTOR substrates identified to date. -

Supplementary Information Material and Methods
MCT-11-0474 BKM120: a potent and specific pan-PI3K inhibitor Supplementary Information Material and methods Chemicals The EGFR inhibitor NVP-AEE788 (Novartis), the Jak inhibitor I (Merck Calbiochem, #420099) and anisomycin (Alomone labs, # A-520) were prepared as 50 mM stock solutions in 100% DMSO. Doxorubicin (Adriablastin, Pfizer), EGF (Sigma Ref: E9644), PDGF (Sigma, Ref: P4306) and IL-4 (Sigma, Ref: I-4269) stock solutions were prepared as recommended by the manufacturer. For in vivo administration: Temodal (20 mg Temozolomide capsules, Essex Chemie AG, Luzern) was dissolved in 4 mL KZI/glucose (20/80, vol/vol); Taxotere was bought as 40 mg/mL solution (Sanofi Aventis, France), and prepared in KZI/glucose. Antibodies The primary antibodies used were as follows: anti-S473P-Akt (#9271), anti-T308P-Akt (#9276,), anti-S9P-GSK3β (#9336), anti-T389P-p70S6K (#9205), anti-YP/TP-Erk1/2 (#9101), anti-YP/TP-p38 (#9215), anti-YP/TP-JNK1/2 (#9101), anti-Y751P-PDGFR (#3161), anti- p21Cip1/Waf1 (#2946), anti-p27Kip1 (#2552) and anti-Ser15-p53 (#9284) antibodies were from Cell Signaling Technologies; anti-Akt (#05-591), anti-T32P-FKHRL1 (#06-952) and anti- PDGFR (#06-495) antibodies were from Upstate; anti-IGF-1R (#SC-713) and anti-EGFR (#SC-03) antibodies were from Santa Cruz; anti-GSK3α/β (#44610), anti-Y641P-Stat6 (#611566), anti-S1981P-ATM (#200-301), anti-T2609 DNA-PKcs (#GTX24194) and anti- 1 MCT-11-0474 BKM120: a potent and specific pan-PI3K inhibitor Y1316P-IGF-1R were from Bio-Source International, Becton-Dickinson, Rockland, GenTex and internal production, respectively. The 4G10 antibody was from Millipore (#05-321MG). -

Genome-Wide Association Study to Identify Genomic Regions And
www.nature.com/scientificreports OPEN Genome‑wide association study to identify genomic regions and positional candidate genes associated with male fertility in beef cattle H. Sweett1, P. A. S. Fonseca1, A. Suárez‑Vega1, A. Livernois1,2, F. Miglior1 & A. Cánovas1* Fertility plays a key role in the success of calf production, but there is evidence that reproductive efciency in beef cattle has decreased during the past half‑century worldwide. Therefore, identifying animals with superior fertility could signifcantly impact cow‑calf production efciency. The objective of this research was to identify candidate regions afecting bull fertility in beef cattle and positional candidate genes annotated within these regions. A GWAS using a weighted single‑step genomic BLUP approach was performed on 265 crossbred beef bulls to identify markers associated with scrotal circumference (SC) and sperm motility (SM). Eight windows containing 32 positional candidate genes and fve windows containing 28 positional candidate genes explained more than 1% of the genetic variance for SC and SM, respectively. These windows were selected to perform gene annotation, QTL enrichment, and functional analyses. Functional candidate gene prioritization analysis revealed 14 prioritized candidate genes for SC of which MAP3K1 and VIP were previously found to play roles in male fertility. A diferent set of 14 prioritized genes were identifed for SM and fve were previously identifed as regulators of male fertility (SOD2, TCP1, PACRG, SPEF2, PRLR). Signifcant enrichment results were identifed for fertility and body conformation QTLs within the candidate windows. Gene ontology enrichment analysis including biological processes, molecular functions, and cellular components revealed signifcant GO terms associated with male fertility. -
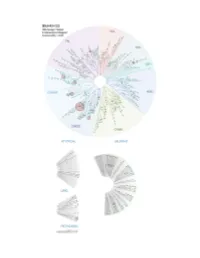
Profiling Data
Compound Name DiscoveRx Gene Symbol Entrez Gene Percent Compound Symbol Control Concentration (nM) BSJ-03-123 AAK1 AAK1 94 1000 BSJ-03-123 ABL1(E255K)-phosphorylated ABL1 79 1000 BSJ-03-123 ABL1(F317I)-nonphosphorylated ABL1 89 1000 BSJ-03-123 ABL1(F317I)-phosphorylated ABL1 98 1000 BSJ-03-123 ABL1(F317L)-nonphosphorylated ABL1 86 1000 BSJ-03-123 ABL1(F317L)-phosphorylated ABL1 89 1000 BSJ-03-123 ABL1(H396P)-nonphosphorylated ABL1 76 1000 BSJ-03-123 ABL1(H396P)-phosphorylated ABL1 90 1000 BSJ-03-123 ABL1(M351T)-phosphorylated ABL1 100 1000 BSJ-03-123 ABL1(Q252H)-nonphosphorylated ABL1 56 1000 BSJ-03-123 ABL1(Q252H)-phosphorylated ABL1 97 1000 BSJ-03-123 ABL1(T315I)-nonphosphorylated ABL1 100 1000 BSJ-03-123 ABL1(T315I)-phosphorylated ABL1 85 1000 BSJ-03-123 ABL1(Y253F)-phosphorylated ABL1 100 1000 BSJ-03-123 ABL1-nonphosphorylated ABL1 60 1000 BSJ-03-123 ABL1-phosphorylated ABL1 79 1000 BSJ-03-123 ABL2 ABL2 89 1000 BSJ-03-123 ACVR1 ACVR1 100 1000 BSJ-03-123 ACVR1B ACVR1B 95 1000 BSJ-03-123 ACVR2A ACVR2A 100 1000 BSJ-03-123 ACVR2B ACVR2B 96 1000 BSJ-03-123 ACVRL1 ACVRL1 84 1000 BSJ-03-123 ADCK3 CABC1 90 1000 BSJ-03-123 ADCK4 ADCK4 91 1000 BSJ-03-123 AKT1 AKT1 100 1000 BSJ-03-123 AKT2 AKT2 98 1000 BSJ-03-123 AKT3 AKT3 100 1000 BSJ-03-123 ALK ALK 100 1000 BSJ-03-123 ALK(C1156Y) ALK 78 1000 BSJ-03-123 ALK(L1196M) ALK 100 1000 BSJ-03-123 AMPK-alpha1 PRKAA1 93 1000 BSJ-03-123 AMPK-alpha2 PRKAA2 100 1000 BSJ-03-123 ANKK1 ANKK1 89 1000 BSJ-03-123 ARK5 NUAK1 98 1000 BSJ-03-123 ASK1 MAP3K5 100 1000 BSJ-03-123 ASK2 MAP3K6 92 1000 BSJ-03-123 AURKA -

Profiling Data
Compound Name DiscoveRx Gene Symbol Entrez Gene Percent Compound Symbol Control Concentration (nM) JNK-IN-8 AAK1 AAK1 69 1000 JNK-IN-8 ABL1(E255K)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317I)-nonphosphorylated ABL1 87 1000 JNK-IN-8 ABL1(F317I)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317L)-nonphosphorylated ABL1 65 1000 JNK-IN-8 ABL1(F317L)-phosphorylated ABL1 61 1000 JNK-IN-8 ABL1(H396P)-nonphosphorylated ABL1 42 1000 JNK-IN-8 ABL1(H396P)-phosphorylated ABL1 60 1000 JNK-IN-8 ABL1(M351T)-phosphorylated ABL1 81 1000 JNK-IN-8 ABL1(Q252H)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(Q252H)-phosphorylated ABL1 56 1000 JNK-IN-8 ABL1(T315I)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(T315I)-phosphorylated ABL1 92 1000 JNK-IN-8 ABL1(Y253F)-phosphorylated ABL1 71 1000 JNK-IN-8 ABL1-nonphosphorylated ABL1 97 1000 JNK-IN-8 ABL1-phosphorylated ABL1 100 1000 JNK-IN-8 ABL2 ABL2 97 1000 JNK-IN-8 ACVR1 ACVR1 100 1000 JNK-IN-8 ACVR1B ACVR1B 88 1000 JNK-IN-8 ACVR2A ACVR2A 100 1000 JNK-IN-8 ACVR2B ACVR2B 100 1000 JNK-IN-8 ACVRL1 ACVRL1 96 1000 JNK-IN-8 ADCK3 CABC1 100 1000 JNK-IN-8 ADCK4 ADCK4 93 1000 JNK-IN-8 AKT1 AKT1 100 1000 JNK-IN-8 AKT2 AKT2 100 1000 JNK-IN-8 AKT3 AKT3 100 1000 JNK-IN-8 ALK ALK 85 1000 JNK-IN-8 AMPK-alpha1 PRKAA1 100 1000 JNK-IN-8 AMPK-alpha2 PRKAA2 84 1000 JNK-IN-8 ANKK1 ANKK1 75 1000 JNK-IN-8 ARK5 NUAK1 100 1000 JNK-IN-8 ASK1 MAP3K5 100 1000 JNK-IN-8 ASK2 MAP3K6 93 1000 JNK-IN-8 AURKA AURKA 100 1000 JNK-IN-8 AURKA AURKA 84 1000 JNK-IN-8 AURKB AURKB 83 1000 JNK-IN-8 AURKB AURKB 96 1000 JNK-IN-8 AURKC AURKC 95 1000 JNK-IN-8 -

PDF Download
MEK-7 Polyclonal Antibody Catalog No : YT2725 Reactivity : Human,Mouse,Rat Applications : WB,ELISA Gene Name : MAP2K7 Protein Name : Dual specificity mitogen-activated protein kinase kinase 7 Human Gene Id : 5609 Human Swiss Prot O14733 No : Mouse Gene Id : 26400 Mouse Swiss Prot Q8CE90 No : Rat Gene Id : 363855 Rat Swiss Prot No : Q4KSH7 Immunogen : The antiserum was produced against synthesized peptide derived from human MAP2K7. AA range:241-290 Specificity : MEK-7 Polyclonal Antibody detects endogenous levels of MEK-7 protein. Formulation : Liquid in PBS containing 50% glycerol, 0.5% BSA and 0.02% sodium azide. Source : Rabbit Dilution : Western Blot: 1/500 - 1/2000. ELISA: 1/20000. Not yet tested in other applications. Purification : The antibody was affinity-purified from rabbit antiserum by affinity- chromatography using epitope-specific immunogen. Concentration : 1 mg/ml 1 / 3 Storage Stability : -20°C/1 year Molecularweight : 47485 Observed Band : 47 Cell Pathway : MAPK_ERK_Growth,MAPK_G_Protein,ErbB_HER,Toll_Like,T_Cell_Receptor, Fc epsilon RI,Neurotrophin,GnRH, Background : mitogen-activated protein kinase kinase 7(MAP2K7) Homo sapiens The protein encoded by this gene is a dual specificity protein kinase that belongs to the MAP kinase kinase family. This kinase specifically activates MAPK8/JNK1 and MAPK9/JNK2, and this kinase itself is phosphorylated and activated by MAP kinase kinase kinases including MAP3K1/MEKK1, MAP3K2/MEKK2,MAP3K3/MEKK5, and MAP4K2/GCK. This kinase is involved in the signal transduction mediating the cell -

Application of a MYC Degradation
SCIENCE SIGNALING | RESEARCH ARTICLE CANCER Copyright © 2019 The Authors, some rights reserved; Application of a MYC degradation screen identifies exclusive licensee American Association sensitivity to CDK9 inhibitors in KRAS-mutant for the Advancement of Science. No claim pancreatic cancer to original U.S. Devon R. Blake1, Angelina V. Vaseva2, Richard G. Hodge2, McKenzie P. Kline3, Thomas S. K. Gilbert1,4, Government Works Vikas Tyagi5, Daowei Huang5, Gabrielle C. Whiten5, Jacob E. Larson5, Xiaodong Wang2,5, Kenneth H. Pearce5, Laura E. Herring1,4, Lee M. Graves1,2,4, Stephen V. Frye2,5, Michael J. Emanuele1,2, Adrienne D. Cox1,2,6, Channing J. Der1,2* Stabilization of the MYC oncoprotein by KRAS signaling critically promotes the growth of pancreatic ductal adeno- carcinoma (PDAC). Thus, understanding how MYC protein stability is regulated may lead to effective therapies. Here, we used a previously developed, flow cytometry–based assay that screened a library of >800 protein kinase inhibitors and identified compounds that promoted either the stability or degradation of MYC in a KRAS-mutant PDAC cell line. We validated compounds that stabilized or destabilized MYC and then focused on one compound, Downloaded from UNC10112785, that induced the substantial loss of MYC protein in both two-dimensional (2D) and 3D cell cultures. We determined that this compound is a potent CDK9 inhibitor with a previously uncharacterized scaffold, caused MYC loss through both transcriptional and posttranslational mechanisms, and suppresses PDAC anchorage- dependent and anchorage-independent growth. We discovered that CDK9 enhanced MYC protein stability 62 through a previously unknown, KRAS-independent mechanism involving direct phosphorylation of MYC at Ser .