DBC1 Does Not Function As a Negative Regulator of SIRT1 in Liver Cancer
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Exploring Prostate Cancer Genome Reveals Simultaneous Losses of PTEN, FAS and PAPSS2 in Patients with PSA Recurrence After Radical Prostatectomy
Int. J. Mol. Sci. 2015, 16, 3856-3869; doi:10.3390/ijms16023856 OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Article Exploring Prostate Cancer Genome Reveals Simultaneous Losses of PTEN, FAS and PAPSS2 in Patients with PSA Recurrence after Radical Prostatectomy Chinyere Ibeawuchi 1, Hartmut Schmidt 2, Reinhard Voss 3, Ulf Titze 4, Mahmoud Abbas 5, Joerg Neumann 6, Elke Eltze 7, Agnes Marije Hoogland 8, Guido Jenster 9, Burkhard Brandt 10 and Axel Semjonow 1,* 1 Prostate Center, Department of Urology, University Hospital Muenster, Albert-Schweitzer-Campus 1, Gebaeude 1A, Muenster D-48149, Germany; E-Mail: [email protected] 2 Center for Laboratory Medicine, University Hospital Muenster, Albert-Schweitzer-Campus 1, Gebaeude 1A, Muenster D-48149, Germany; E-Mail: [email protected] 3 Interdisciplinary Center for Clinical Research, University of Muenster, Albert-Schweitzer-Campus 1, Gebaeude D3, Domagkstrasse 3, Muenster D-48149, Germany; E-Mail: [email protected] 4 Pathology, Lippe Hospital Detmold, Röntgenstrasse 18, Detmold D-32756, Germany; E-Mail: [email protected] 5 Institute of Pathology, Mathias-Spital-Rheine, Frankenburg Street 31, Rheine D-48431, Germany; E-Mail: [email protected] 6 Institute of Pathology, Klinikum Osnabrueck, Am Finkenhuegel 1, Osnabrueck D-49076, Germany; E-Mail: [email protected] 7 Institute of Pathology, Saarbrücken-Rastpfuhl, Rheinstrasse 2, Saarbrücken D-66113, Germany; E-Mail: [email protected] 8 Department -

Ten Commandments for a Good Scientist
Unravelling the mechanism of differential biological responses induced by food-borne xeno- and phyto-estrogenic compounds Ana María Sotoca Covaleda Wageningen 2010 Thesis committee Thesis supervisors Prof. dr. ir. Ivonne M.C.M. Rietjens Professor of Toxicology Wageningen University Prof. dr. Albertinka J. Murk Personal chair at the sub-department of Toxicology Wageningen University Thesis co-supervisor Dr. ir. Jacques J.M. Vervoort Associate professor at the Laboratory of Biochemistry Wageningen University Other members Prof. dr. Michael R. Muller, Wageningen University Prof. dr. ir. Huub F.J. Savelkoul, Wageningen University Prof. dr. Everardus J. van Zoelen, Radboud University Nijmegen Dr. ir. Toine F.H. Bovee, RIKILT, Wageningen This research was conducted under the auspices of the Graduate School VLAG Unravelling the mechanism of differential biological responses induced by food-borne xeno- and phyto-estrogenic compounds Ana María Sotoca Covaleda Thesis submitted in fulfillment of the requirements for the degree of doctor at Wageningen University by the authority of the Rector Magnificus Prof. dr. M.J. Kropff, in the presence of the Thesis Committee appointed by the Academic Board to be defended in public on Tuesday 14 September 2010 at 4 p.m. in the Aula Unravelling the mechanism of differential biological responses induced by food-borne xeno- and phyto-estrogenic compounds. Ana María Sotoca Covaleda Thesis Wageningen University, Wageningen, The Netherlands, 2010, With references, and with summary in Dutch. ISBN: 978-90-8585-707-5 “Caminante no hay camino, se hace camino al andar. Al andar se hace camino, y al volver la vista atrás se ve la senda que nunca se ha de volver a pisar” - Antonio Machado – A mi madre. -

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -

Understanding the Role of Perilipin 5 in Non-Alcoholic Fatty Liver Disease and Its Role in Hepatocellular Carcinoma: a Review of Novel Insights
International Journal of Molecular Sciences Review Understanding the Role of Perilipin 5 in Non-Alcoholic Fatty Liver Disease and Its Role in Hepatocellular Carcinoma: A Review of Novel Insights Paola Berenice Mass Sanchez, Marinela Krizanac, Ralf Weiskirchen * and Anastasia Asimakopoulos * Institute of Molecular Pathobiochemistry, Experimental Gene Therapy and Clinical Chemistry (IFMPEGKC), RWTH University Hospital Aachen, D-52074 Aachen, Germany; [email protected] (P.B.M.S.); [email protected] (M.K.) * Correspondence: [email protected] (R.W.); [email protected] (A.A.) Abstract: Consumption of high-calorie foods, such as diets rich in fats, is an important factor leading to the development of steatohepatitis. Several studies have suggested how lipid accumulation creates a lipotoxic microenvironment for cells, leading cells to deregulate their transcriptional and transla- tional activity. This deregulation induces the development of liver diseases such as non-alcoholic fatty liver disease (NAFLD) and subsequently also the appearance of hepatocellular carcinoma (HCC) which is one of the deadliest types of cancers worldwide. Understanding its pathology and studying new biomarkers with better specificity in predicting disease prognosis can help in the personalized treatment of the disease. In this setting, understanding the link between NAFLD and HCC progression, the differentiation of each stage in between as well as the mechanisms underlying this process, are vital for development of new treatments and in exploring new therapeutic targets. Citation: Mass Sanchez, P.B.; Perilipins are a family of five closely related proteins expressed on the surface of lipid droplets (LD) in Krizanac, M.; Weiskirchen, R.; several tissues acting in several pathways involved in lipid metabolism. -

A Dissertation Entitled the Androgen Receptor
A Dissertation entitled The Androgen Receptor as a Transcriptional Co-activator: Implications in the Growth and Progression of Prostate Cancer By Mesfin Gonit Submitted to the Graduate Faculty as partial fulfillment of the requirements for the PhD Degree in Biomedical science Dr. Manohar Ratnam, Committee Chair Dr. Lirim Shemshedini, Committee Member Dr. Robert Trumbly, Committee Member Dr. Edwin Sanchez, Committee Member Dr. Beata Lecka -Czernik, Committee Member Dr. Patricia R. Komuniecki, Dean College of Graduate Studies The University of Toledo August 2011 Copyright 2011, Mesfin Gonit This document is copyrighted material. Under copyright law, no parts of this document may be reproduced without the expressed permission of the author. An Abstract of The Androgen Receptor as a Transcriptional Co-activator: Implications in the Growth and Progression of Prostate Cancer By Mesfin Gonit As partial fulfillment of the requirements for the PhD Degree in Biomedical science The University of Toledo August 2011 Prostate cancer depends on the androgen receptor (AR) for growth and survival even in the absence of androgen. In the classical models of gene activation by AR, ligand activated AR signals through binding to the androgen response elements (AREs) in the target gene promoter/enhancer. In the present study the role of AREs in the androgen- independent transcriptional signaling was investigated using LP50 cells, derived from parental LNCaP cells through extended passage in vitro. LP50 cells reflected the signature gene overexpression profile of advanced clinical prostate tumors. The growth of LP50 cells was profoundly dependent on nuclear localized AR but was independent of androgen. Nevertheless, in these cells AR was unable to bind to AREs in the absence of androgen. -

Delineation of Key Regulatory Elements Identifies Points Of
DELINEATION OF KEY REGULATORY ELEMENTS IDENTIFIES POINTS OF VULNERABILITY IN THE MITOGEN-ACTIVATED SIGNALING NETWORK SUPPLEMENTARY MATERIALS List of contents Supplementary Figures with legends 1. Figure S1: Distribution of primary siRNA screen data, and standardization of assay procedure. 2. Figure S2: Scatter plot of screen data. 3. Figure S3: Functional relevance of the identified targets and Calculation of residence time from PDT and cell cycle distribution. 4. Figure S4: FACS profiles for ABL1 and AKT1. Table for data in Figure 5B. 5. Figure S5: Venn diagram showing the results of the comparative analysis of other screen results 6. Figure S6: Dose response profiles for the AKT1 + ABL1 inhibitor combination for CH1, list of the 14 cell lines and their description, effect of ABL1+AKT1 inhibitor combination on increase in apoptotic cells and G1 arrest in 14 cell lines, effects of CHEK1 inhibitor on combination C1,C2 on 4 cell lines. Supplementary Tables 1. Table S1: siRNA screen results for targeted kinases and phosphatases. 2. Table S2: Gene expression status of the validated hits. 3. Table S3: Role played by identified RNAi hits in regulation of cell cycle, the effect on PDTs along with phase-specific RTs. 4. Table S4: List of molecules classified as cell cycle targets. 5. Table S5: High confidence network used for graph theory analysis. 6. Table S6: Occurrences of nodes in shortest path networks. 7. Table S7: Network file used as SNAVI background. 8. Table S8: Classification of nodes present in modules according to specificity. Legends for tables Supplementary Experimental Procedures References Figure S1 A 450 400 G1 S 350 G2 300 250 200 150 100 50 Distribution of molecules Distribution 0 -6-4-20246 Z-score 350 200 400 G1 S 300 G2 150 300 250 200 100 200 150 100 50 100 Distribution of molecules 50 0 0 0 -4 -2 0 2 4 -4-20246 -4-20246 Z-score B PLK1 GAPDH PLCg BTK PLCg CDC2A PLCg CHEK1 PLCg MET Distribution profiles of complete primary screen and western blots showing knockdown efficiency. -

Characterisation of Isomirs in Stem Cells
Characterisation of isomiRs in stem cells Geok Chin Tan Institute of Reproductive and Developmental Biology Department of Surgery and Cancer Faculty of Medicine Imperial College London Thesis submitted to Imperial College London for the degree of Doctor of Philosophy 1 Statement of Originality All experiments included in this thesis were performed by me unless otherwise stated in the text. 2 Copyright Statement ‘The copyright of this thesis rests with the author and is made available under a Creative Commons Attribution Non-Commercial No Derivatives licence. Researchers are free to copy, distribute or transmit the thesis on the condition that they attribute it, that they do not use it for commercial purposes and that they do not alter, transform or build upon it. For any reuse or redistribution, researchers must make clear to others the licence terms of this work’ 3 Acknowledgements I would like to thank my supervisor Dr Nicholas Dibb for giving me the opportunity to work in his lab and for all of his guidance and support throughout my PhD, without which this project would not have been possible. I am also very grateful to Dr Wei Cui for teaching me the technique of stem cell culture, her comments on my project related to stem cells and as a wonderful co-supervisor. I would like to also thank Professor Malcolm Parker for his supports and advise on academic and non-academic related subjects. Many thanks to Elcie Chan for the generation of all the stem cell libraries which forms the platform for my project. My sincere thanks also to Gunter Meister for supplying the Argonaute antibodies, Leandro Castellano for the help in the design of RNA sponges, Laki Buluwela for the pTRIPz lentiviral vector and last but not least Alywn Dart from Charlotte Bevan group for the prostate cancer cell lines. -
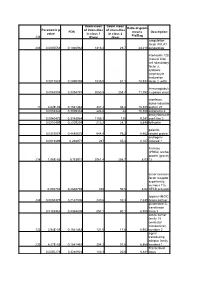
238 Parametric P- Value FDR Geom Mean of Intensities in Class 1 (Plate
Geom mean Geom mean Ratio of geom Parametric p- of intensities of intensities FDR means Description value in class 1 in class 2 Pla/Bag 238 (Plate) (Bag) coagulation factor XIII, A1 206 0,0005558 0,1940962 1313,3 29,7 44,219 polypeptide interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation 0,0017883 0,2490398 1535,5 81,1 18,933 factor 2, p40) Immunoglobuli 0,0063804 0,3394787 3050,5 258,7 11,792 n epsilon chain interferon, alpha-inducible 11 4,62E-05 0,1541482 347,2 33,4 10,395 protein 27 0,0035346 0,3039332 226,8 22 10,309 astrotactin 2 deoxyribonucle 0,0040473 0,3148984 1155,1 125 9,241 ase I-like 3 0,0010495 0,2205294 213,2 24,1 8,846 follistatin galectin- 0,0315078 0,4885675 644,8 76,2 8,462 related protein androgen- 0,0013899 0,238971 287 35,4 8,107 induced 1 A kinase (PRKA) anchor protein (gravin) 236 1,06E-05 0,115911 2061,4 256,7 8,03 12 tumor necrosis factor receptor superfamily, member 11a, 0,002746 0,2846759 405 50,5 8,02 NFKB activator lipoma HMGIC 248 0,0008375 0,2147006 240,6 32,3 7,449 fusion partner glutathione S- transferase 0,0183462 0,4368809 557,1 80,1 6,955 theta 1 solute carrier family 18 (vesicular monoamine), 122 3,94E-05 0,1541482 121,5 17,6 6,903 member 2 signal transducing adaptor family 233 6,27E-05 0,1541482 354,3 51,6 6,866 member 1 Transcribed 0,0050276 0,3240923 140,3 20,6 6,811 locus aldehyde dehydrogenas e 5 family, member A1 (succinate- semialdehyde dehydrogenas 0,0294865 0,4819829 236,6 38,2 6,194 e) guanylate cyclase activator 1A 0,0074099 0,3504479 339,1 55 6,165 -

Identification of 5 Novel Genes Methylated in Breast and Other Epithelial Cancers
Hill et al. Molecular Cancer 2010, 9:51 http://www.molecular-cancer.com/content/9/1/51 RESEARCH Open Access Identification of 5 novel genes methylated in breast and other epithelial cancers Victoria K Hill1, Luke B Hesson1,2, Temuujin Dansranjavin3, Ashraf Dallol1, Ivan Bieche4, Sophie Vacher4, Stella Tommasi5, Timothy Dobbins2, Dean Gentle1, David Euhus6, Cheryl Lewis6, Reinhard Dammann3, Robyn L Ward2, John Minna7, Eammon R Maher1, Gerd P Pfeifer5, Farida Latif1* Abstract Background: There are several high throughput approaches to identify methylated genes in cancer. We utilized one such recently developed approach, MIRA (methylated-CpG island recovery assay) combined with CpG island arrays to identify novel genes that are epigenetically inactivated in breast cancer. Results: Using this approach we identified numerous CpG islands that demonstrated aberrant DNA methylation in breast cancer cell lines. Using a combination of COBRA and sequencing of bisulphite modified DNA, we confirmed 5 novel genes frequently methylated in breast tumours; EMILIN2, SALL1, DBC1, FBLN2 and CIDE-A. Methylation frequencies ranged from between 25% and 63% in primary breast tumours, whilst matched normal breast tissue DNA was either unmethylated or demonstrated a much lower frequency of methylation compared to malignant breast tissue DNA. Furthermore expression of the above 5 genes was shown to be restored following treatment with a demethylating agent in methylated breast cancer cell lines. We have expanded this analysis across three other common epithelial cancers (lung, colorectal, prostate). We demonstrate that the above genes show varying levels of methylation in these cancers. Lastly and most importantly methylation of EMILIN2 was associated with poorer clinical outcome in breast cancer and was strongly associated with estrogen receptor as well as progesterone receptor positive breast cancers. -

Transdifferentiation of Human Mesenchymal Stem Cells
Transdifferentiation of Human Mesenchymal Stem Cells Dissertation zur Erlangung des naturwissenschaftlichen Doktorgrades der Julius-Maximilians-Universität Würzburg vorgelegt von Tatjana Schilling aus San Miguel de Tucuman, Argentinien Würzburg, 2007 Eingereicht am: Mitglieder der Promotionskommission: Vorsitzender: Prof. Dr. Martin J. Müller Gutachter: PD Dr. Norbert Schütze Gutachter: Prof. Dr. Georg Krohne Tag des Promotionskolloquiums: Doktorurkunde ausgehändigt am: Hiermit erkläre ich ehrenwörtlich, dass ich die vorliegende Dissertation selbstständig angefertigt und keine anderen als die von mir angegebenen Hilfsmittel und Quellen verwendet habe. Des Weiteren erkläre ich, dass diese Arbeit weder in gleicher noch in ähnlicher Form in einem Prüfungsverfahren vorgelegen hat und ich noch keinen Promotionsversuch unternommen habe. Gerbrunn, 4. Mai 2007 Tatjana Schilling Table of contents i Table of contents 1 Summary ........................................................................................................................ 1 1.1 Summary.................................................................................................................... 1 1.2 Zusammenfassung..................................................................................................... 2 2 Introduction.................................................................................................................... 4 2.1 Osteoporosis and the fatty degeneration of the bone marrow..................................... 4 2.2 Adipose and bone -

DBC1 Antibody A
C 0 2 - t DBC1 Antibody a e r o t S Orders: 877-616-CELL (2355) [email protected] Support: 877-678-TECH (8324) 3 9 Web: [email protected] 6 www.cellsignal.com 5 # 3 Trask Lane Danvers Massachusetts 01923 USA For Research Use Only. Not For Use In Diagnostic Procedures. Applications: Reactivity: Sensitivity: MW (kDa): Source: UniProt ID: Entrez-Gene Id: WB, IP, IF-IC H M R Mk Endogenous 130 Rabbit Q8N163 57805 Product Usage Information 1. Hamaguchi, M. et al. (2002) Proc Natl Acad Sci USA 99, 13647-52. 2. Trauernicht, A.M. et al. (2007) Mol Endocrinol 21, 1526-36. Application Dilution 3. Zhao, W. et al. (2008) Nature 451, 587-90. 4. Li, Z. et al. (2009) J Biol Chem 284, 10361-6. Western Blotting 1:1000 5. Sundararajan, R. et al. (2005) Oncogene 24, 4908-20. Immunoprecipitation 1:50 Immunofluorescence (Immunocytochemistry) 1:100 Storage Supplied in 10 mM sodium HEPES (pH 7.5), 150 mM NaCl, 100 µg/ml BSA and 50% glycerol. Store at –20°C. Do not aliquot the antibody. Specificity / Sensitivity DBC1 Antibody detects endogenous levels of total DBC1 protein. Species Reactivity: Human, Mouse, Rat, Monkey Species predicted to react based on 100% sequence homology: Horse Source / Purification Polyclonal antibodies are produced by immunizing animals with a synthetic peptide corresponding to residues near the amino terminus of human DBC1 protein. Antibodies are purified by protein A and peptide affinity chromatography. Background Deleted in breast cancer gene 1 protein (DBC1) was originally identified by its localization to a region of chromosome 8p21 that is homozygously deleted in breast cancer (1). -

Identification of Breast Cancer Subtype-Specific Biomarkers By
medicina Article Identification of Breast Cancer Subtype-Specific Biomarkers by Integrating Copy Number Alterations and Gene Expression Profiles Claudia Cava 1,* , Mirko Pisati 1,2 , Marco Frasca 2 and Isabella Castiglioni 3 1 Institute of Molecular Bioimaging and Physiology, National Research Council (IBFM-CNR), Via F. Cervi 93, Segrate-Milan, 20090 Milan, Italy; [email protected] 2 Department of Computer Science, Università degli Studi di Milano, Via Celoria 18, 20133 Milano, Italy; [email protected] 3 Department of Physics “Giuseppe Occhialini”, University of Milan-Bicocca Piazza dell’Ateneo Nuovo, 20126 Milan, Italy; [email protected] * Correspondence: [email protected] Abstract: Background and Objectives: Breast cancer is a heterogeneous disease categorized into four subtypes. Previous studies have shown that copy number alterations of several genes are implicated with the development and progression of many cancers. This study evaluates the effects of DNA copy number alterations on gene expression levels in different breast cancer subtypes. Materials and Methods: We performed a computational analysis integrating copy number alterations and gene expression profiles in 1024 breast cancer samples grouped into four molecular subtypes: luminal A, luminal B, HER2, and basal. Results: Our analyses identified several genes correlated in all Citation: Cava, C.; Pisati, M.; Frasca, subtypes such as KIAA1967 and MCPH1. In addition, several subtype-specific genes that showed a M.; Castiglioni, I. Identification of significant correlation between copy number and gene expression profiles were detected: SMARCB1, Breast Cancer Subtype-Specific AZIN1, MTDH in luminal A, PPP2R5E, APEX1, GCN5 in luminal B, TNFAIP1, PCYT2, DIABLO Biomarkers by Integrating Copy in HER2, and FAM175B, SENP5, SCAF1 in basal subtype.