The Study of Copy Number Variations in the Regions of PRKAB2 and PPM1K Among Congenital Heart Defects Patients
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplemental Information to Mammadova-Bach Et Al., “Laminin Α1 Orchestrates VEGFA Functions in the Ecosystem of Colorectal Carcinogenesis”
Supplemental information to Mammadova-Bach et al., “Laminin α1 orchestrates VEGFA functions in the ecosystem of colorectal carcinogenesis” Supplemental material and methods Cloning of the villin-LMα1 vector The plasmid pBS-villin-promoter containing the 3.5 Kb of the murine villin promoter, the first non coding exon, 5.5 kb of the first intron and 15 nucleotides of the second villin exon, was generated by S. Robine (Institut Curie, Paris, France). The EcoRI site in the multi cloning site was destroyed by fill in ligation with T4 polymerase according to the manufacturer`s instructions (New England Biolabs, Ozyme, Saint Quentin en Yvelines, France). Site directed mutagenesis (GeneEditor in vitro Site-Directed Mutagenesis system, Promega, Charbonnières-les-Bains, France) was then used to introduce a BsiWI site before the start codon of the villin coding sequence using the 5’ phosphorylated primer: 5’CCTTCTCCTCTAGGCTCGCGTACGATGACGTCGGACTTGCGG3’. A double strand annealed oligonucleotide, 5’GGCCGGACGCGTGAATTCGTCGACGC3’ and 5’GGCCGCGTCGACGAATTCACGC GTCC3’ containing restriction site for MluI, EcoRI and SalI were inserted in the NotI site (present in the multi cloning site), generating the plasmid pBS-villin-promoter-MES. The SV40 polyA region of the pEGFP plasmid (Clontech, Ozyme, Saint Quentin Yvelines, France) was amplified by PCR using primers 5’GGCGCCTCTAGATCATAATCAGCCATA3’ and 5’GGCGCCCTTAAGATACATTGATGAGTT3’ before subcloning into the pGEMTeasy vector (Promega, Charbonnières-les-Bains, France). After EcoRI digestion, the SV40 polyA fragment was purified with the NucleoSpin Extract II kit (Machery-Nagel, Hoerdt, France) and then subcloned into the EcoRI site of the plasmid pBS-villin-promoter-MES. Site directed mutagenesis was used to introduce a BsiWI site (5’ phosphorylated AGCGCAGGGAGCGGCGGCCGTACGATGCGCGGCAGCGGCACG3’) before the initiation codon and a MluI site (5’ phosphorylated 1 CCCGGGCCTGAGCCCTAAACGCGTGCCAGCCTCTGCCCTTGG3’) after the stop codon in the full length cDNA coding for the mouse LMα1 in the pCIS vector (kindly provided by P. -

Brief Genetics Report Haplotype Structures and Large
Brief Genetics Report Haplotype Structures and Large-Scale Association Testing of the 5 AMP-Activated Protein Kinase Genes PRKAA2, PRKAB1, and PRKAB2 With Type 2 Diabetes Maria W. Sun,1,2 Jennifer Y. Lee,1,2 Paul I.W. de Bakker,1,2,3 Noe¨l P. Burtt,2 Peter Almgren,4 Lennart Råstam,5 Tiinamaija Tuomi,6 Daniel Gaudet,7 Mark J. Daly,2,8 Joel N. Hirschhorn,2,3,9 David Altshuler,1,2,3,8,10 Leif Groop,4,6 and Jose C. Florez1,2,8,10 AMP-activated protein kinase (AMPK) is a key molecular plasma glucose, or insulin sensitivity. Several nominal asso- regulator of cellular metabolism, and its activity is induced ciations of variants in PRKAA2 and PRKAB1 with BMI appear by both metformin and thiazolidinedione antidiabetic med- to be consistent with statistical noise. Diabetes 55:849–855, ications. It has therefore been proposed both as a putative 2006 agent in the pathophysiology of type 2 diabetes and as a valid target for therapeutic intervention. Thus, the genes that encode the various AMPK subunits are intriguing ype 2 diabetes arises from the complex interplay candidates for the inherited basis of type 2 diabetes. We therefore set out to test for the association of common of various pathophysiologic mechanisms involv- variants in the genes that encode three selected AMPK ing peripheral insulin resistance and relative subunits with type 2 diabetes and related phenotypes. Of Tinsulin insufficiency. The final expression of the the seven genes that encode AMPK isoforms, we initially diabetic phenotype is strongly influenced by inheritance; chose PRKAA2, PRKAB1, and PRKAB2 because of their however, with the exception of rare monogenic forms of higher prior probability of association with type 2 diabetes, diabetes, common type 2 diabetes is thought to have a based on previous reports of genetic linkage, functional polygenic architecture (1). -

Transcriptomic Characterization of Fibrolamellar Hepatocellular
Transcriptomic characterization of fibrolamellar PNAS PLUS hepatocellular carcinoma Elana P. Simona, Catherine A. Freijeb, Benjamin A. Farbera,c, Gadi Lalazara, David G. Darcya,c, Joshua N. Honeymana,c, Rachel Chiaroni-Clarkea, Brian D. Dilld, Henrik Molinad, Umesh K. Bhanote, Michael P. La Quagliac, Brad R. Rosenbergb,f, and Sanford M. Simona,1 aLaboratory of Cellular Biophysics, The Rockefeller University, New York, NY 10065; bPresidential Fellows Laboratory, The Rockefeller University, New York, NY 10065; cDivision of Pediatric Surgery, Department of Surgery, Memorial Sloan-Kettering Cancer Center, New York, NY 10065; dProteomics Resource Center, The Rockefeller University, New York, NY 10065; ePathology Core Facility, Memorial Sloan-Kettering Cancer Center, New York, NY 10065; and fJohn C. Whitehead Presidential Fellows Program, The Rockefeller University, New York, NY 10065 Edited by Susan S. Taylor, University of California, San Diego, La Jolla, CA, and approved September 22, 2015 (received for review December 29, 2014) Fibrolamellar hepatocellular carcinoma (FLHCC) tumors all carry a exon of DNAJB1 and all but the first exon of PRKACA. This deletion of ∼400 kb in chromosome 19, resulting in a fusion of the produced a chimeric RNA transcript and a translated chimeric genes for the heat shock protein, DNAJ (Hsp40) homolog, subfam- protein that retains the full catalytic activity of wild-type PKA. ily B, member 1, DNAJB1, and the catalytic subunit of protein ki- This chimeric protein was found in 15 of 15 FLHCC patients nase A, PRKACA. The resulting chimeric transcript produces a (21) in the absence of any other recurrent mutations in the DNA fusion protein that retains kinase activity. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Age, DNA Methylation and the Malignant Potential of the Serrated Neoplasia Pathway Lochlan John Fennell B
Age, DNA Methylation and the Malignant Potential of the Serrated Neoplasia Pathway Lochlan John Fennell B. Biomed Sci A thesis submitted for the degree of Doctor of Philosophy at The University of Queensland in 2020 Faculty of Medicine ORC ID: 0000-0003-3214-3527 1 Abstract Colorectal cancer is the third most common cancer in Australia and is responsible for the death of over four thousand Australians each year. There are two overarching molecular pathways leading to colorectal cancer. The conventional pathway, which is responsible for ~75% of colorectal cancer diagnoses, occurs in a step-wise manner and is the consequence of a series of genetic alterations including mutations of tumour suppressor genes and gross chromosomal abnormalities. This pathway has been extensively studied over the past three decades. The serrated neoplasia pathway is responsible for the remaining colorectal cancers. This pathway is triggered by oncogenic BRAF mutation and these cancers accumulate epigenetic alterations while progressing to invasive cancer. DNA methylation is important in serrated neoplasia, however the extent and role of DNA methylation on the initiation and progression of serrated lesions is not clear. DNA methylation accumulates in tissues with age, and advanced serrated lesions and cancers occur almost exclusively in elderly patients. How this methylation affects serrated lesions is unknown. In this thesis I set out to address three key research questions related to DNA methylation, age and serrated colorectal neoplasia. First, what is the extent of DNA methylation in colorectal cancers?; Second, Does age-related hypermethylation, and namely that occurring at the loci encoding tumour suppressor genes, increase the risk of serrated colorectal neoplasia?; and if true, how can we reconcile this with the existence of early onset serrated colorectal cancer? In the first chapter of this thesis, I examine the DNA methylation and transcriptional architecture of 216 colorectal cancer samples collected consecutively at the Royal Brisbane and Women’s hospital. -

A New Computational Approach to Evaluating Systemic Gene–Gene Interactions in a Pathway Affected by Drug LY294002
processes Article A New Computational Approach to Evaluating Systemic Gene–Gene Interactions in a Pathway Affected by Drug LY294002 Shinuk Kim College of Kyedang General Education, Sangmyung University, Cheonan 31066, Korea; [email protected]; Tel.: +82-(41)-550-5452; Fax: +82-(41)-550-5439 Received: 14 August 2020; Accepted: 23 September 2020; Published: 1 October 2020 Abstract: In this study, we investigate how drugs systemically affect genes via pathways by integrating information from interactions between chemical compounds and molecular expression datasets, and from pathway information such as gene sets using mathematical models. First, we adopt drug-induced gene expression datasets; then, employ gene set enrichment analysis tools for selecting candidate enrichment pathways; and lastly, implement the inverse algorithm package for identifying gene–gene regulatory networks in a pathway. We tested LY294002-induced datasets of the MCF7 breast cancer cell lines, and found a CELL CYCLE pathway with 101 genes, ERBB signaling pathway consisting of 82 genes, and MTOR pathway consisting of 45 genes. We consider two interactions: quantity strength depending on number of interactions, and quality strength depending on weight of interaction as positive (+) and negative ( ) interactions. Our methods revealed ANAPC1-CDK6 ( 0.412) and − − ORC2L- CHEK1(0.951) for the CELL CYCLE pathway; INS-RPS6 ( 3.125) and PRKAA2-PRKAA2 − (+1.319) for the MTOR pathway; and CBLB-RPS6KB1 ( 0.141), RPS6KB1-CBLC (+0.238) for the ERBB − signaling pathway to be top quality interactions. Top quantity interactions discovered include 12; the CDC ( ,+) gene family for the CELL CYCLE pathway, 20; PIK3 ( ), 23; PIK3CG (+) for the MTOR − − pathway, 11; PAK ( ), 10; PIK3 (+) for the ERBB signaling pathway. -

Association Study of AMP-Activated Protein Kinase Subunit Genes In
European Journal of Endocrinology (2009) 161 405–409 ISSN 0804-4643 CLINICAL STUDY Association study of AMP-activated protein kinase subunit genes in polycystic ovary syndrome Kari Sproul1,2, Michelle R Jones3, Ricardo Azziz1,2,4 and Mark O Goodarzi1,3,4,5 1Department of Obstetrics and Gynecology, Cedars-Sinai Medical Center, Los Angeles, California 90048, USA, 2Department of Obstetrics and Gynecology, the David Geffen School of Medicine at UCLA, Los Angeles, California 90095, USA, 3Division of Endocrinology, Diabetes and Metabolism, Department of Medicine, Cedars-Sinai Medical Center, 8700 Beverly Boulevard, Room B-131, Los Angeles, California 90048, USA, 4Department of Medicine, the David Geffen School of Medicine at UCLA, Los Angeles, California 90095, USA and 5Medical Genetics Institute, Cedars-Sinai Medical Center, Los Angeles, California 90048, USA (Correspondence should be addressed to M O Goodarzi at Division of Endocrinology, Diabetes and Metabolism, Department of Medicine, Cedars-Sinai Medical Center; Email: [email protected]) Abstract Objective: To examine the genes for AMP-activated protein kinase (AMPK) subunits a2(PRKAA2) and g3(PRKAG3) as candidates for polycystic ovary syndrome (PCOS) and its component traits. Design and methods: A total of 287 white PCOS women were recruited from the reproductive endocrinology clinic at the University of Alabama at Birmingham and 187 white control subjects were recruited from the surrounding community. Seven PRKAA2 single nucleotide polymorphisms (SNPs) and four PRKAG3 SNPs were genotyped in PCOS cases and controls. Genotyping and association analysis were performed at Cedars-Sinai Medical Center. Results: Nominal associations of PRKAA2 variants with insulin-related traits and the PRKAG3 Pro71Ala variant with PCOS were not statistically significant after multiple testing correction. -

Ahn Supp. Fig. 1 AB 1.5 ARRDC4 1.5 ARRDC4 * * * 1.0 1.0
Ahn_Supp. Fig. 1 AB 1.5 ARRDC4 1.5 ARRDC4 * * * 1.0 1.0 * * 0.5 * 0.5 * * * Relative mRNA levels mRNA Relative Relative mRNA levels mRNA Relative 0.0 0.0 1.5 MLXIP (MondoA) 1.5 MLXIP (MondoA) 1.0 1.0 0.5 0.5 Relative mRNA levels mRNA Relative Relative mRNA levels mRNA Relative 0.0 0.0 MondoA MondoA 0124824 Starvation (6h) -++++++ Glucose Starvation (h) Refeeding (h) --0.51248 C 1.5 ARRDC4 1.5 MLXIP (MondoA) † Con # KD 1.0 1.0 0.5 0.5 * * * * Relative mRNA levels mRNA Relative Relative mRNA levels mRNA Relative * * 0.0 0.0 BasalStarvation Refeeding BasalStarvation Refeeding MondoA Con + + - - + + - - + + - - KD - - + + - - + + - - + + BasalStarvation Refeeding Supplemental Figure 1. Glucose-mediated regulation of ARRDC4 is dependent on MondoA in human skeletal myotubes. (A) (top) ARRDC4 and MLXIP (MondoA) mRNA levels were determined by qRT-PCR in human skeletal myotubes following deprivation of glucose at the indicated time (n=4). (bottom) Representative Western blot analysis of MondoA demonstrating the effect of glucose deprivation. *p<0.05 vs. 0h. (B) (top) ARRDC4 and MLXIP (MondoA) expression in human myotubes following a 6h glucose removal and refeeding at the times indicated (n=4). (bottom) Corresponding Western blot analysis. *p<0.05 vs Starvation 6h. (C) (top) Expression of ARRDC4 and MLXIP in human myotubes following deprivation and refeeding of glucose in the absence or presence of siRNA-mediated MondoA KD (n=4). (bottom) Corresponding Western blot analysis. *p<0.05 vs siControl. # p<0.05. § p<0.05. The data represents mean ± SD. All statistical significance determined by one-way ANOVA with Tukey multiple comparison post-hoc test. -
![AMPK Beta 2 (PRKAB2) Mouse Monoclonal Antibody [Clone ID: OTI4H4] Product Data](https://docslib.b-cdn.net/cover/7127/ampk-beta-2-prkab2-mouse-monoclonal-antibody-clone-id-oti4h4-product-data-1047127.webp)
AMPK Beta 2 (PRKAB2) Mouse Monoclonal Antibody [Clone ID: OTI4H4] Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TA808333 AMPK beta 2 (PRKAB2) Mouse Monoclonal Antibody [Clone ID: OTI4H4] Product data: Product Type: Primary Antibodies Clone Name: OTI4H4 Applications: WB Recommended Dilution: WB 1:2000 Reactivity: Human, Mouse, Rat Host: Mouse Isotype: IgG1 Clonality: Monoclonal Immunogen: Full length human recombinant protein of human PRKAB2 (NP_005390) produced in E.coli. Formulation: PBS (PH 7.3) containing 1% BSA, 50% glycerol and 0.02% sodium azide. Concentration: 1 mg/ml Purification: Purified from mouse ascites fluids or tissue culture supernatant by affinity chromatography (protein A/G) Conjugation: Unconjugated Storage: Store at -20°C as received. Stability: Stable for 12 months from date of receipt. Predicted Protein Size: 30.1 kDa Gene Name: protein kinase AMP-activated non-catalytic subunit beta 2 Database Link: NP_005390 Entrez Gene 64562 RatEntrez Gene 5565 Human O43741 This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 2 AMPK beta 2 (PRKAB2) Mouse Monoclonal Antibody [Clone ID: OTI4H4] – TA808333 Background: The protein encoded by this gene is a regulatory subunit of the AMP-activated protein kinase (AMPK). AMPK is a heterotrimer consisting of an alpha catalytic subunit, and non-catalytic beta and gamma subunits. AMPK is an important energy-sensing enzyme that monitors cellular energy status. In response to cellular metabolic stresses, AMPK is activated, and thus phosphorylates and inactivates acetyl-CoA carboxylase (ACC) and beta-hydroxy beta- methylglutaryl-CoA reductase (HMGCR), key enzymes involved in regulating de novo biosynthesis of fatty acid and cholesterol. -
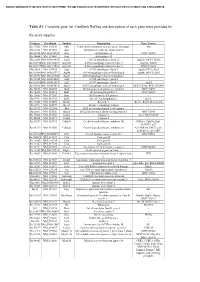
Table S1. Complete Gene List. Genbank Refseq and Description of Each Gene Were Provided By
Document downloaded from http://www.elsevier.es, day 24/09/2021. This copy is for personal use. Any transmission of this document by any media or format is strictly prohibited. Table S1. Complete gene list. GenBank RefSeq and description of each gene were provided by the array supplier. Unigene GeneBank Symbol Description Gene Name/s Rn.11422 NM_033230 Akt1 V-akt murine thymoma viral oncogene homolog 1 Akt Rn.2104 NM_019288 App Amyloid beta (A4) precursor protein - Rn.23323 NM_001034933 Arsa Arylsulfatase A MGC125207 Rn.94004 NM_033443 Arsb Arylsulfatase B - Rn.6224 NM_001038495 Atg12 ATG12 autophagy related 12 Apg12l, MGC125080 Rn.101734NM_001108809 Atg16l1 ATG16 autophagy related 16-like 1 Apg16l, Wdr30 Rn.104199NM_001191560 Atg16l2 ATG16 autophagy related 16-like 2 RGD1311400 Rn.3084 NM_134394 Atg3 ATG3 autophagy related 3 Apg3l, PIG-1, Pig1 Rn.163086NM_001025711 Atg4b ATG4 autophagy related 4 homolog B Apg4b, MGC112887 Rn.23378 NM_001107948 Atg4c ATG4 autophagy related 4 homolog C - Rn.98385 NM_001014250 Atg5 ATG5 autophagy related 5 - Rn.162765NM_001012097 Atg7 ATG7 autophagy related 7 Apg7l Rn.35248 NM_001014218 Atg9a ATG9 autophagy related 9 homolog A MGC105908, RGD1310450 Rn.36696 NM_022698 Bad BCL2-associated agonist of cell death MGC72439 Rn.14598 NM_053812 Bak1 BCL2-antagonist/killer 1 MGC108627 Rn.10668 NM_017059 Bax Bcl2-associated X protein - Rn.9996 NM_016993 Bcl2 B-cell CLL/lymphoma 2 Bcl-2 Rn.10323 NM_031535 Bcl2l1 Bcl2-like 1 Bcl-xl, Bcl2l, Bclx, bcl-X Rn.2776 NM_053739 Becn1 Beclin 1, autophagy related - Rn.31142 NM_022684 -

Development and Validation of a Protein-Based Risk Score for Cardiovascular Outcomes Among Patients with Stable Coronary Heart Disease
Supplementary Online Content Ganz P, Heidecker B, Hveem K, et al. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA. doi: 10.1001/jama.2016.5951 eTable 1. List of 1130 Proteins Measured by Somalogic’s Modified Aptamer-Based Proteomic Assay eTable 2. Coefficients for Weibull Recalibration Model Applied to 9-Protein Model eFigure 1. Median Protein Levels in Derivation and Validation Cohort eTable 3. Coefficients for the Recalibration Model Applied to Refit Framingham eFigure 2. Calibration Plots for the Refit Framingham Model eTable 4. List of 200 Proteins Associated With the Risk of MI, Stroke, Heart Failure, and Death eFigure 3. Hazard Ratios of Lasso Selected Proteins for Primary End Point of MI, Stroke, Heart Failure, and Death eFigure 4. 9-Protein Prognostic Model Hazard Ratios Adjusted for Framingham Variables eFigure 5. 9-Protein Risk Scores by Event Type This supplementary material has been provided by the authors to give readers additional information about their work. Downloaded From: https://jamanetwork.com/ on 10/02/2021 Supplemental Material Table of Contents 1 Study Design and Data Processing ......................................................................................................... 3 2 Table of 1130 Proteins Measured .......................................................................................................... 4 3 Variable Selection and Statistical Modeling ........................................................................................ -

(AMP)-Activated Protein Kinase: a New Target for Nutraceutical Compounds
International Journal of Molecular Sciences Review Adenosine Monophosphate (AMP)-Activated Protein Kinase: A New Target for Nutraceutical Compounds Fabiola Marín-Aguilar 1, Luis E. Pavillard 1, Francesca Giampieri 2, Pedro Bullón 1 and Mario D. Cordero 1,* 1 Research Laboratory, Oral Medicine Department, University of Sevilla, Sevilla 41009, Spain; [email protected] (F.M.-A.); [email protected] (L.E.P.); [email protected] (P.B.) 2 Dipartimento di Scienze Cliniche Specialistiche ed Odontostomatologiche—Sez. Biochimica, Università Politecnica delle Marche, Ancona 60100, Italy; [email protected] * Correspondence: [email protected]; Tel.: +34-954-481-120; Fax: +34-954-486-784 Academic Editor: Rosa M. Lamuela-Raventós Received: 17 November 2016; Accepted: 23 January 2017; Published: 29 January 2017 Abstract: Adenosine monophosphate-activated protein kinase (AMPK) is an important energy sensor which is activated by increases in adenosine monophosphate (AMP)/adenosine triphosphate (ATP) ratio and/or adenosine diphosphate (ADP)/ATP ratio, and increases different metabolic pathways such as fatty acid oxidation, glucose transport and mitochondrial biogenesis. In this sense, AMPK maintains cellular energy homeostasis by induction of catabolism and inhibition of ATP-consuming biosynthetic pathways to preserve ATP levels. Several studies indicate a reduction of AMPK sensitivity to cellular stress during aging and this could impair the downstream signaling and the maintenance of the cellular energy balance and the stress resistance. However, several diseases have been related with an AMPK dysfunction. Alterations in AMPK signaling decrease mitochondrial biogenesis, increase cellular stress and induce inflammation, which are typical events of the aging process and have been associated to several pathological processes.