Identifying Lineage Relationships in Human T Cell Populations
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Table 2. Functional Classification of Genes Differentially Regulated After HOXB4 Inactivation in HSC/Hpcs
Table 2. Functional classification of genes differentially regulated after HOXB4 inactivation in HSC/HPCs Symbol Gene description Fold-change (mean ± SD) Signal transduction Adam8 A disintegrin and metalloprotease domain 8 1.91 ± 0.51 Arl4 ADP-ribosylation factor-like 4 - 1.80 ± 0.40 Dusp6 Dual specificity phosphatase 6 (Mkp3) - 2.30 ± 0.46 Ksr1 Kinase suppressor of ras 1 1.92 ± 0.42 Lyst Lysosomal trafficking regulator 1.89 ± 0.34 Mapk1ip1 Mitogen activated protein kinase 1 interacting protein 1 1.84 ± 0.22 Narf* Nuclear prelamin A recognition factor 2.12 ± 0.04 Plekha2 Pleckstrin homology domain-containing. family A. (phosphoinosite 2.15 ± 0.22 binding specific) member 2 Ptp4a2 Protein tyrosine phosphatase 4a2 - 2.04 ± 0.94 Rasa2* RAS p21 activator protein 2 - 2.80 ± 0.13 Rassf4 RAS association (RalGDS/AF-6) domain family 4 3.44 ± 2.56 Rgs18 Regulator of G-protein signaling - 1.93 ± 0.57 Rrad Ras-related associated with diabetes 1.81 ± 0.73 Sh3kbp1 SH3 domain kinase bindings protein 1 - 2.19 ± 0.53 Senp2 SUMO/sentrin specific protease 2 - 1.97 ± 0.49 Socs2 Suppressor of cytokine signaling 2 - 2.82 ± 0.85 Socs5 Suppressor of cytokine signaling 5 2.13 ± 0.08 Socs6 Suppressor of cytokine signaling 6 - 2.18 ± 0.38 Spry1 Sprouty 1 - 2.69 ± 0.19 Sos1 Son of sevenless homolog 1 (Drosophila) 2.16 ± 0.71 Ywhag 3-monooxygenase/tryptophan 5- monooxygenase activation protein. - 2.37 ± 1.42 gamma polypeptide Zfyve21 Zinc finger. FYVE domain containing 21 1.93 ± 0.57 Ligands and receptors Bambi BMP and activin membrane-bound inhibitor - 2.94 ± 0.62 -

Genomic Correlates of Relationship QTL Involved in Fore- Versus Hind Limb Divergence in Mice
Loyola University Chicago Loyola eCommons Biology: Faculty Publications and Other Works Faculty Publications 2013 Genomic Correlates of Relationship QTL Involved in Fore- Versus Hind Limb Divergence in Mice Mihaela Palicev Gunter P. Wagner James P. Noonan Benedikt Hallgrimsson James M. Cheverud Loyola University Chicago, [email protected] Follow this and additional works at: https://ecommons.luc.edu/biology_facpubs Part of the Biology Commons Recommended Citation Palicev, M, GP Wagner, JP Noonan, B Hallgrimsson, and JM Cheverud. "Genomic Correlates of Relationship QTL Involved in Fore- Versus Hind Limb Divergence in Mice." Genome Biology and Evolution 5(10), 2013. This Article is brought to you for free and open access by the Faculty Publications at Loyola eCommons. It has been accepted for inclusion in Biology: Faculty Publications and Other Works by an authorized administrator of Loyola eCommons. For more information, please contact [email protected]. This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 3.0 License. © Palicev et al., 2013. GBE Genomic Correlates of Relationship QTL Involved in Fore- versus Hind Limb Divergence in Mice Mihaela Pavlicev1,2,*, Gu¨ nter P. Wagner3, James P. Noonan4, Benedikt Hallgrı´msson5,and James M. Cheverud6 1Konrad Lorenz Institute for Evolution and Cognition Research, Altenberg, Austria 2Department of Pediatrics, Cincinnati Children‘s Hospital Medical Center, Cincinnati, Ohio 3Yale Systems Biology Institute and Department of Ecology and Evolutionary Biology, Yale University 4Department of Genetics, Yale University School of Medicine 5Department of Cell Biology and Anatomy, The McCaig Institute for Bone and Joint Health and the Alberta Children’s Hospital Research Institute for Child and Maternal Health, University of Calgary, Calgary, Canada 6Department of Anatomy and Neurobiology, Washington University *Corresponding author: E-mail: [email protected]. -

A Conserved Gene Family Encodes Transmembrane Proteins with Fibronectin, Immunoglobulin and Leucine-Rich Repeat Domains (FIGLER) Delicia L
A conserved gene family encodes transmembrane proteins with fibronectin, immunoglobulin and leucine-rich repeat domains (FIGLER) Delicia L. Munfus, University of Alabama at Birmingham Christopher L. Haga, University of Alabama at Birmingham Peter D. Burrows, University of Alabama at Birmingham Max Cooper, Emory University Journal Title: BMC Biology Volume: Volume 5, Number 1 Publisher: BioMed Central | 2007-09-13, Pages 36-36 Type of Work: Article | Final Publisher PDF Publisher DOI: 10.1186/1741-7007-5-36 Permanent URL: https://pid.emory.edu/ark:/25593/rqdhh Final published version: http://dx.doi.org/10.1186/1741-7007-5-36 Copyright information: © 2007 Munfus et al; licensee BioMed Central Ltd. This is an Open Access work distributed under the terms of the Creative Commons Attribution 2.0 Generic License (http://creativecommons.org/licenses/by/2.0/). Accessed October 6, 2021 6:19 AM EDT BMC Biology BioMed Central Research article Open Access A conserved gene family encodes transmembrane proteins with fibronectin, immunoglobulin and leucine-rich repeat domains (FIGLER) Delicia L Munfus†1,2, Christopher L Haga†1,2, Peter D Burrows1,2,3 and Max D Cooper*1,2,4,5 Address: 1Division of Developmental and Clinical Immunology, University of Alabama at Birmingham, Birmingham, AL 35294-3300, USA, 2Department of Microbiology, University of Alabama at Birmingham, Birmingham, AL 35294-3300, USA, 3Department of Genetics, University of Alabama at Birmingham, Birmingham, AL 35294-3300,, 4Department of Medicine, University of Alabama at Birmingham, Birmingham, -

Supp Table 1.Pdf
Upregulated genes in Hdac8 null cranial neural crest cells fold change Gene Symbol Gene Title 134.39 Stmn4 stathmin-like 4 46.05 Lhx1 LIM homeobox protein 1 31.45 Lect2 leukocyte cell-derived chemotaxin 2 31.09 Zfp108 zinc finger protein 108 27.74 0710007G10Rik RIKEN cDNA 0710007G10 gene 26.31 1700019O17Rik RIKEN cDNA 1700019O17 gene 25.72 Cyb561 Cytochrome b-561 25.35 Tsc22d1 TSC22 domain family, member 1 25.27 4921513I08Rik RIKEN cDNA 4921513I08 gene 24.58 Ofa oncofetal antigen 24.47 B230112I24Rik RIKEN cDNA B230112I24 gene 23.86 Uty ubiquitously transcribed tetratricopeptide repeat gene, Y chromosome 22.84 D8Ertd268e DNA segment, Chr 8, ERATO Doi 268, expressed 19.78 Dag1 Dystroglycan 1 19.74 Pkn1 protein kinase N1 18.64 Cts8 cathepsin 8 18.23 1500012D20Rik RIKEN cDNA 1500012D20 gene 18.09 Slc43a2 solute carrier family 43, member 2 17.17 Pcm1 Pericentriolar material 1 17.17 Prg2 proteoglycan 2, bone marrow 17.11 LOC671579 hypothetical protein LOC671579 17.11 Slco1a5 solute carrier organic anion transporter family, member 1a5 17.02 Fbxl7 F-box and leucine-rich repeat protein 7 17.02 Kcns2 K+ voltage-gated channel, subfamily S, 2 16.93 AW493845 Expressed sequence AW493845 16.12 1600014K23Rik RIKEN cDNA 1600014K23 gene 15.71 Cst8 cystatin 8 (cystatin-related epididymal spermatogenic) 15.68 4922502D21Rik RIKEN cDNA 4922502D21 gene 15.32 2810011L19Rik RIKEN cDNA 2810011L19 gene 15.08 Btbd9 BTB (POZ) domain containing 9 14.77 Hoxa11os homeo box A11, opposite strand transcript 14.74 Obp1a odorant binding protein Ia 14.72 ORF28 open reading -

A B-Cell Receptor-Related Gene Signature Predicts Survival in Mantle
Published Ahead of Print on February 22, 2018, as doi:10.3324/haematol.2017.184325. Copyright 2018 Ferrata Storti Foundation. A B-cell receptor-related gene signature predicts survival in mantle cell lymphoma: results from the “Fondazione Italiana Linfomi” MCL-0208 trial by Riccardo Bomben, Simone Ferrero, Tiziana D'Agaro, Michele Dal Bo, Alessandro Re, Andrea Evangelista, Angelo Michele Carella, Alberto Zamò, Umberto Vitolo, Paola Omedè, Chiara Rusconi, Luca Arcaini, Luigi Rigacci, Stefano Luminari, Andrea Piccin, Delong Liu, Adrien Wiestner, Gianluca Gaidano, Sergio Cortelazzo, Marco Ladetto, and Valter Gattei Haematologica 2018 [Epub ahead of print] Citation: Riccardo Bomben, Simone Ferrero, Tiziana D'Agaro, Michele Dal Bo, Alessandro Re, Andrea Evangelista, Angelo Michele Carella, Alberto Zamò, Umberto Vitolo, Paola Omedè, Chiara Rusconi, Luca Arcaini, Luigi Rigacci, Stefano Luminari, Andrea Piccin, Delong Liu, Adrien Wiestner, Gianluca Gaidano, Sergio Cortelazzo, Marco Ladetto, and Valter Gattei. A B-cell receptor-related gene signature predicts survival in mantle cell lymphoma: results from the “Fondazione Italiana Linfomi” MCL-0208 trial. Haematologica. 2018; 103:xxx doi:10.3324/haematol.2017.184325 Publisher's Disclaimer. E-publishing ahead of print is increasingly important for the rapid dissemination of science. Haematologica is, therefore, E-publishing PDF files of an early version of manuscripts that have completed a regular peer review and have been accepted for publication. E-publishing of this PDF file has been approved by the authors. After having E-published Ahead of Print, manuscripts will then undergo technical and English editing, typesetting, proof correction and be presented for the authors' final approval; the final version of the manuscript will then appear in print on a regular issue of the journal. -

Genetic Drivers of Pancreatic Islet Function
| INVESTIGATION Genetic Drivers of Pancreatic Islet Function Mark P. Keller,*,1 Daniel M. Gatti,†,1 Kathryn L. Schueler,* Mary E. Rabaglia,* Donnie S. Stapleton,* Petr Simecek,† Matthew Vincent,† Sadie Allen,‡ Aimee Teo Broman,§ Rhonda Bacher,§ Christina Kendziorski,§ Karl W. Broman,§ Brian S. Yandell,** Gary A. Churchill,†,2 and Alan D. Attie*,2 *Department of Biochemistry, §Department of Biostatistics and Medical Informatics, and **Department of Horticulture, University of Wisconsin–Madison, Wisconsin 53706-1544, †The Jackson Laboratory, Bar Harbor, Maine 06409, and ‡Maine School of Science and Mathematics, Limestone, Maine 06409, ORCID IDs: 0000-0002-7405-5552 (M.P.K.); 0000-0002-4914-6671 (K.W.B.); 0000-0001-9190-9284 (G.A.C.); 0000-0002-0568-2261 (A.D.A.) ABSTRACT The majority of gene loci that have been associated with type 2 diabetes play a role in pancreatic islet function. To evaluate the role of islet gene expression in the etiology of diabetes, we sensitized a genetically diverse mouse population with a Western diet high in fat (45% kcal) and sucrose (34%) and carried out genome-wide association mapping of diabetes-related phenotypes. We quantified mRNA abundance in the islets and identified 18,820 expression QTL. We applied mediation analysis to identify candidate causal driver genes at loci that affect the abundance of numerous transcripts. These include two genes previously associated with monogenic diabetes (PDX1 and HNF4A), as well as three genes with nominal association with diabetes-related traits in humans (FAM83E, IL6ST, and SAT2). We grouped transcripts into gene modules and mapped regulatory loci for modules enriched with transcripts specific for a-cells, and another specific for d-cells. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -
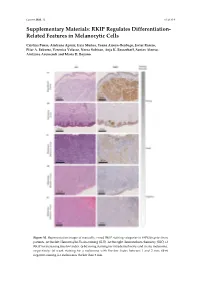
RKIP Regulates Differentiation- Related Features in Melanocytic Cells
Cancers 2020. 12 S1 of S14 Supplementary Materials: RKIP Regulates Differentiation- Related Features in Melanocytic Cells Cristina Penas, Aintzane Apraiz, Iraia Muñoa, Yoana Arroyo-Berdugo, Javier Rasero, Pilar A. Ezkurra, Veronica Velasco, Nerea Subiran, Anja K. Bosserhoff, Santos Alonso, Aintzane Asumendi and Maria D. Boyano Figure S1. Representative images of manually scored RKIP staining categories in FFPE biopsies from patients. At the feft: Hematoxylin-Eosin staining (H-E). At the right: Immunohistochemistry (IHC) of RKIP for increasing Breslow Index. (a-b) Strong staining for intradermal nevus and in situ melanoma, respectively; (c) weak staining for a melanoma with Breslow Index between 1 and 2 mm; (d-e) negative staining for melanomas thicker than 2 mm. Cancers 2020. 12 S2 of S14 Figure S2. RNA Sequencing data analysis. Each row of the figure represents an over-represented gene signature generated by analyzing the up-regulated and down-regulated genes from a differential expression analysis of RKIP Knockdown melanocytes HEMn-LP. Results from the EnrichR Website with a p-value > 0.05. Figure S3. RNA Sequencing data quality. (a) Read counts normalization per sample; (b) Spearman correlation coefficient between replicates; (c) Principal component analysis for replicates. Cancers 2020. 12 S3 of S14 Figure S4. Details of Western Blot from Figure 2 related with RKIP expression in several cell lines. (a) Blots for RKIP and Tubulin in primary and metastatic melanoma and in three primary melanocytes HEMn-DP (dark pigmented), HEMn-LP (light pigmented) and HEM2710. On the right, original blot for each analyzed protein. Highlighted with a red square the information included in the Figure 3 (horizontal rotation). -

Neural Development
31 October 2007 NEURAL DEVELOPMENT www.neuraldevelopment.com Analysis of Lrrn1 expression and its relationship to neuromeric boundaries during chick neural development Laura C Andreae et al. Neural Development 2007, 2:22 http://www.neuraldevelopment.com/content/2/1/22 Neural Development BioMed Central Research article Open Access Analysis of Lrrn1 expression and its relationship to neuromeric boundaries during chick neural development LauraCAndreae1,2, Daniela Peukert1, Andrew Lumsden1 and Jonathan D Gilthorpe*1 Address: 1MRC Centre for Developmental Neurobiology, King's College London, New Hunt's House, Guy's Campus, London, UK, SE1 1UL and 2Department of Neurophysiology, National Institute for Medical Research, The Ridgeway, Mill Hill, London, UK, NW7 1AA Email: Laura C Andreae - [email protected]; Daniela Peukert - [email protected]; Andrew Lumsden - [email protected]; Jonathan D Gilthorpe* - [email protected] * Corresponding author Published: 31 October 2007 Received: 26 March 2007 Accepted: 31 October 2007 Neural Development 2007, 2:22 doi:10.1186/1749-8104-2-22 This article is available from: http://www.neuraldevelopment.com/content/2/1/22 © 2007 Andreae et al.; licensee BioMed Central Ltd. This is an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: The Drosophila leucine-rich repeat proteins Tartan (TRN) and Capricious (CAPS) mediate cell affinity differences during compartition of the wing imaginal disc. This study aims to identify and characterize the expression of a chick orthologue of TRN/CAPS and examine its potential function in relation to compartment boundaries in the vertebrate central nervous system. -

View / Download 3.3 Mb
Identification of Mechanisms and Pathways Involved in MLL2-Mediated Tumorigenesis by Chun-Chi Chang Department of Pathology Duke University Date:_______________________ Approved: ___________________________ Yiping He, Supervisor ___________________________ Salvatore Pizzo ___________________________ Hai Yan Thesis submitted in partial fulfillment of the requirements for the degree of Master of Science in the Department of Pathology in the Graduate School of Duke University 2013 ABSTRACT Identification of Mechanisms and Pathways Involved in MLL2-Mediated Tumorigenesis by Chun-Chi Chang Department of Pathology Duke University Date:_______________________ Approved: ___________________________ Yiping He, Supervisor ___________________________ Salvatore Pizzo ___________________________ Hai Yan An abstract of a thesis submitted in partial fulfillment of the requirements for the degree of Master of Science in the Department of Pathology in the Graduate School of Duke University 2013 Copyright by Chun-Chi Chang 2013 Abstract Myeloid/lymphoid or mixed-lineage leukemia (MLL)-family genes encode histone lysine methyltransferases that play important roles in epigenetic regulation of gene transcription, and these genes are frequently mutated in human cancers. While MLL1 and MLL4 have been the most extensively studied, MLL2 and its homolog MLL3 are not well-understood. Specifically, little is known regarding the extent of global MLL2 involvement in the regulation of gene expression and the mechanism underlying its alterations in mediating tumorigenesis. To study the role of MLL2 in tumorigenesis, we somatically knocked out MLL2 in a colorectal carcinoma cell line, HCT116. We observed that the MLL2 loss of function results in significant reduction of cell growth and multinuclear morphology. We further profiled MLL2 regulated genes and pathways by analyzing gene expression in MLL2 wild-type versus MLL2-null isogenic cell lines. -

Supp Table 6.Pdf
Supplementary Table 6. Processes associated to the 2037 SCL candidate target genes ID Symbol Entrez Gene Name Process NM_178114 AMIGO2 adhesion molecule with Ig-like domain 2 adhesion NM_033474 ARVCF armadillo repeat gene deletes in velocardiofacial syndrome adhesion NM_027060 BTBD9 BTB (POZ) domain containing 9 adhesion NM_001039149 CD226 CD226 molecule adhesion NM_010581 CD47 CD47 molecule adhesion NM_023370 CDH23 cadherin-like 23 adhesion NM_207298 CERCAM cerebral endothelial cell adhesion molecule adhesion NM_021719 CLDN15 claudin 15 adhesion NM_009902 CLDN3 claudin 3 adhesion NM_008779 CNTN3 contactin 3 (plasmacytoma associated) adhesion NM_015734 COL5A1 collagen, type V, alpha 1 adhesion NM_007803 CTTN cortactin adhesion NM_009142 CX3CL1 chemokine (C-X3-C motif) ligand 1 adhesion NM_031174 DSCAM Down syndrome cell adhesion molecule adhesion NM_145158 EMILIN2 elastin microfibril interfacer 2 adhesion NM_001081286 FAT1 FAT tumor suppressor homolog 1 (Drosophila) adhesion NM_001080814 FAT3 FAT tumor suppressor homolog 3 (Drosophila) adhesion NM_153795 FERMT3 fermitin family homolog 3 (Drosophila) adhesion NM_010494 ICAM2 intercellular adhesion molecule 2 adhesion NM_023892 ICAM4 (includes EG:3386) intercellular adhesion molecule 4 (Landsteiner-Wiener blood group)adhesion NM_001001979 MEGF10 multiple EGF-like-domains 10 adhesion NM_172522 MEGF11 multiple EGF-like-domains 11 adhesion NM_010739 MUC13 mucin 13, cell surface associated adhesion NM_013610 NINJ1 ninjurin 1 adhesion NM_016718 NINJ2 ninjurin 2 adhesion NM_172932 NLGN3 neuroligin -

Supplemental Table S1. Primers for Sybrgreen Quantitative RT-PCR Assays
Supplemental Table S1. Primers for SYBRGreen quantitative RT-PCR assays. Gene Accession Primer Sequence Length Start Stop Tm GC% GAPDH NM_002046.3 GAPDH F TCCTGTTCGACAGTCAGCCGCA 22 39 60 60.43 59.09 GAPDH R GCGCCCAATACGACCAAATCCGT 23 150 128 60.12 56.52 Exon junction 131/132 (reverse primer) on template NM_002046.3 DNAH6 NM_001370.1 DNAH6 F GGGCCTGGTGCTGCTTTGATGA 22 4690 4711 59.66 59.09% DNAH6 R TAGAGAGCTTTGCCGCTTTGGCG 23 4797 4775 60.06 56.52% Exon junction 4790/4791 (reverse primer) on template NM_001370.1 DNAH7 NM_018897.2 DNAH7 F TGCTGCATGAGCGGGCGATTA 21 9973 9993 59.25 57.14% DNAH7 R AGGAAGCCATGTACAAAGGTTGGCA 25 10073 10049 58.85 48.00% Exon junction 9989/9990 (forward primer) on template NM_018897.2 DNAI1 NM_012144.2 DNAI1 F AACAGATGTGCCTGCAGCTGGG 22 673 694 59.67 59.09 DNAI1 R TCTCGATCCCGGACAGGGTTGT 22 822 801 59.07 59.09 Exon junction 814/815 (reverse primer) on template NM_012144.2 RPGRIP1L NM_015272.2 RPGRIP1L F TCCCAAGGTTTCACAAGAAGGCAGT 25 3118 3142 58.5 48.00% RPGRIP1L R TGCCAAGCTTTGTTCTGCAAGCTGA 25 3238 3214 60.06 48.00% Exon junction 3124/3125 (forward primer) on template NM_015272.2 Supplemental Table S2. Transcripts that differentiate IPF/UIP from controls at 5%FDR Fold- p-value Change Transcript Gene p-value p-value p-value (IPF/UIP (IPF/UIP Cluster ID RefSeq Symbol gene_assignment (Age) (Gender) (Smoking) vs. C) vs. C) NM_001178008 // CBS // cystathionine-beta- 8070632 NM_001178008 CBS synthase // 21q22.3 // 875 /// NM_0000 0.456642 0.314761 0.418564 4.83E-36 -2.23 NM_003013 // SFRP2 // secreted frizzled- 8103254 NM_003013