A Novel LRR and Ig Domain-Containing Protein Could Function As an Immune Effector in Crassostrea Gigas T
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Table 2. Functional Classification of Genes Differentially Regulated After HOXB4 Inactivation in HSC/Hpcs
Table 2. Functional classification of genes differentially regulated after HOXB4 inactivation in HSC/HPCs Symbol Gene description Fold-change (mean ± SD) Signal transduction Adam8 A disintegrin and metalloprotease domain 8 1.91 ± 0.51 Arl4 ADP-ribosylation factor-like 4 - 1.80 ± 0.40 Dusp6 Dual specificity phosphatase 6 (Mkp3) - 2.30 ± 0.46 Ksr1 Kinase suppressor of ras 1 1.92 ± 0.42 Lyst Lysosomal trafficking regulator 1.89 ± 0.34 Mapk1ip1 Mitogen activated protein kinase 1 interacting protein 1 1.84 ± 0.22 Narf* Nuclear prelamin A recognition factor 2.12 ± 0.04 Plekha2 Pleckstrin homology domain-containing. family A. (phosphoinosite 2.15 ± 0.22 binding specific) member 2 Ptp4a2 Protein tyrosine phosphatase 4a2 - 2.04 ± 0.94 Rasa2* RAS p21 activator protein 2 - 2.80 ± 0.13 Rassf4 RAS association (RalGDS/AF-6) domain family 4 3.44 ± 2.56 Rgs18 Regulator of G-protein signaling - 1.93 ± 0.57 Rrad Ras-related associated with diabetes 1.81 ± 0.73 Sh3kbp1 SH3 domain kinase bindings protein 1 - 2.19 ± 0.53 Senp2 SUMO/sentrin specific protease 2 - 1.97 ± 0.49 Socs2 Suppressor of cytokine signaling 2 - 2.82 ± 0.85 Socs5 Suppressor of cytokine signaling 5 2.13 ± 0.08 Socs6 Suppressor of cytokine signaling 6 - 2.18 ± 0.38 Spry1 Sprouty 1 - 2.69 ± 0.19 Sos1 Son of sevenless homolog 1 (Drosophila) 2.16 ± 0.71 Ywhag 3-monooxygenase/tryptophan 5- monooxygenase activation protein. - 2.37 ± 1.42 gamma polypeptide Zfyve21 Zinc finger. FYVE domain containing 21 1.93 ± 0.57 Ligands and receptors Bambi BMP and activin membrane-bound inhibitor - 2.94 ± 0.62 -

Genomic Correlates of Relationship QTL Involved in Fore- Versus Hind Limb Divergence in Mice
Loyola University Chicago Loyola eCommons Biology: Faculty Publications and Other Works Faculty Publications 2013 Genomic Correlates of Relationship QTL Involved in Fore- Versus Hind Limb Divergence in Mice Mihaela Palicev Gunter P. Wagner James P. Noonan Benedikt Hallgrimsson James M. Cheverud Loyola University Chicago, [email protected] Follow this and additional works at: https://ecommons.luc.edu/biology_facpubs Part of the Biology Commons Recommended Citation Palicev, M, GP Wagner, JP Noonan, B Hallgrimsson, and JM Cheverud. "Genomic Correlates of Relationship QTL Involved in Fore- Versus Hind Limb Divergence in Mice." Genome Biology and Evolution 5(10), 2013. This Article is brought to you for free and open access by the Faculty Publications at Loyola eCommons. It has been accepted for inclusion in Biology: Faculty Publications and Other Works by an authorized administrator of Loyola eCommons. For more information, please contact [email protected]. This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 3.0 License. © Palicev et al., 2013. GBE Genomic Correlates of Relationship QTL Involved in Fore- versus Hind Limb Divergence in Mice Mihaela Pavlicev1,2,*, Gu¨ nter P. Wagner3, James P. Noonan4, Benedikt Hallgrı´msson5,and James M. Cheverud6 1Konrad Lorenz Institute for Evolution and Cognition Research, Altenberg, Austria 2Department of Pediatrics, Cincinnati Children‘s Hospital Medical Center, Cincinnati, Ohio 3Yale Systems Biology Institute and Department of Ecology and Evolutionary Biology, Yale University 4Department of Genetics, Yale University School of Medicine 5Department of Cell Biology and Anatomy, The McCaig Institute for Bone and Joint Health and the Alberta Children’s Hospital Research Institute for Child and Maternal Health, University of Calgary, Calgary, Canada 6Department of Anatomy and Neurobiology, Washington University *Corresponding author: E-mail: [email protected]. -

A Conserved Gene Family Encodes Transmembrane Proteins with Fibronectin, Immunoglobulin and Leucine-Rich Repeat Domains (FIGLER) Delicia L
A conserved gene family encodes transmembrane proteins with fibronectin, immunoglobulin and leucine-rich repeat domains (FIGLER) Delicia L. Munfus, University of Alabama at Birmingham Christopher L. Haga, University of Alabama at Birmingham Peter D. Burrows, University of Alabama at Birmingham Max Cooper, Emory University Journal Title: BMC Biology Volume: Volume 5, Number 1 Publisher: BioMed Central | 2007-09-13, Pages 36-36 Type of Work: Article | Final Publisher PDF Publisher DOI: 10.1186/1741-7007-5-36 Permanent URL: https://pid.emory.edu/ark:/25593/rqdhh Final published version: http://dx.doi.org/10.1186/1741-7007-5-36 Copyright information: © 2007 Munfus et al; licensee BioMed Central Ltd. This is an Open Access work distributed under the terms of the Creative Commons Attribution 2.0 Generic License (http://creativecommons.org/licenses/by/2.0/). Accessed October 6, 2021 6:19 AM EDT BMC Biology BioMed Central Research article Open Access A conserved gene family encodes transmembrane proteins with fibronectin, immunoglobulin and leucine-rich repeat domains (FIGLER) Delicia L Munfus†1,2, Christopher L Haga†1,2, Peter D Burrows1,2,3 and Max D Cooper*1,2,4,5 Address: 1Division of Developmental and Clinical Immunology, University of Alabama at Birmingham, Birmingham, AL 35294-3300, USA, 2Department of Microbiology, University of Alabama at Birmingham, Birmingham, AL 35294-3300, USA, 3Department of Genetics, University of Alabama at Birmingham, Birmingham, AL 35294-3300,, 4Department of Medicine, University of Alabama at Birmingham, Birmingham, -

Genetic Drivers of Pancreatic Islet Function
| INVESTIGATION Genetic Drivers of Pancreatic Islet Function Mark P. Keller,*,1 Daniel M. Gatti,†,1 Kathryn L. Schueler,* Mary E. Rabaglia,* Donnie S. Stapleton,* Petr Simecek,† Matthew Vincent,† Sadie Allen,‡ Aimee Teo Broman,§ Rhonda Bacher,§ Christina Kendziorski,§ Karl W. Broman,§ Brian S. Yandell,** Gary A. Churchill,†,2 and Alan D. Attie*,2 *Department of Biochemistry, §Department of Biostatistics and Medical Informatics, and **Department of Horticulture, University of Wisconsin–Madison, Wisconsin 53706-1544, †The Jackson Laboratory, Bar Harbor, Maine 06409, and ‡Maine School of Science and Mathematics, Limestone, Maine 06409, ORCID IDs: 0000-0002-7405-5552 (M.P.K.); 0000-0002-4914-6671 (K.W.B.); 0000-0001-9190-9284 (G.A.C.); 0000-0002-0568-2261 (A.D.A.) ABSTRACT The majority of gene loci that have been associated with type 2 diabetes play a role in pancreatic islet function. To evaluate the role of islet gene expression in the etiology of diabetes, we sensitized a genetically diverse mouse population with a Western diet high in fat (45% kcal) and sucrose (34%) and carried out genome-wide association mapping of diabetes-related phenotypes. We quantified mRNA abundance in the islets and identified 18,820 expression QTL. We applied mediation analysis to identify candidate causal driver genes at loci that affect the abundance of numerous transcripts. These include two genes previously associated with monogenic diabetes (PDX1 and HNF4A), as well as three genes with nominal association with diabetes-related traits in humans (FAM83E, IL6ST, and SAT2). We grouped transcripts into gene modules and mapped regulatory loci for modules enriched with transcripts specific for a-cells, and another specific for d-cells. -
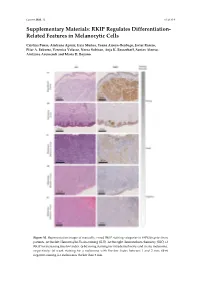
RKIP Regulates Differentiation- Related Features in Melanocytic Cells
Cancers 2020. 12 S1 of S14 Supplementary Materials: RKIP Regulates Differentiation- Related Features in Melanocytic Cells Cristina Penas, Aintzane Apraiz, Iraia Muñoa, Yoana Arroyo-Berdugo, Javier Rasero, Pilar A. Ezkurra, Veronica Velasco, Nerea Subiran, Anja K. Bosserhoff, Santos Alonso, Aintzane Asumendi and Maria D. Boyano Figure S1. Representative images of manually scored RKIP staining categories in FFPE biopsies from patients. At the feft: Hematoxylin-Eosin staining (H-E). At the right: Immunohistochemistry (IHC) of RKIP for increasing Breslow Index. (a-b) Strong staining for intradermal nevus and in situ melanoma, respectively; (c) weak staining for a melanoma with Breslow Index between 1 and 2 mm; (d-e) negative staining for melanomas thicker than 2 mm. Cancers 2020. 12 S2 of S14 Figure S2. RNA Sequencing data analysis. Each row of the figure represents an over-represented gene signature generated by analyzing the up-regulated and down-regulated genes from a differential expression analysis of RKIP Knockdown melanocytes HEMn-LP. Results from the EnrichR Website with a p-value > 0.05. Figure S3. RNA Sequencing data quality. (a) Read counts normalization per sample; (b) Spearman correlation coefficient between replicates; (c) Principal component analysis for replicates. Cancers 2020. 12 S3 of S14 Figure S4. Details of Western Blot from Figure 2 related with RKIP expression in several cell lines. (a) Blots for RKIP and Tubulin in primary and metastatic melanoma and in three primary melanocytes HEMn-DP (dark pigmented), HEMn-LP (light pigmented) and HEM2710. On the right, original blot for each analyzed protein. Highlighted with a red square the information included in the Figure 3 (horizontal rotation). -

Identifying Lineage Relationships in Human T Cell Populations
Identifying lineage relationships in human T cell populations by Celia Lara Menckeberg A thesis submitted to The University of Birmingham for the degree of DOCTOR OF PHILOSOPHY School of Immunity and Infection College of Medical and Dental Sciences The University of Birmingham December 2010 University of Birmingham Research Archive e-theses repository This unpublished thesis/dissertation is copyright of the author and/or third parties. The intellectual property rights of the author or third parties in respect of this work are as defined by The Copyright Designs and Patents Act 1988 or as modified by any successor legislation. Any use made of information contained in this thesis/dissertation must be in accordance with that legislation and must be properly acknowledged. Further distribution or reproduction in any format is prohibited without the permission of the copyright holder. ii ABSTRACT CD4+ and CD8+ T cell populations can be divided into subpopulations based on expression of surface markers CCR7 and CD45RA. The resulting populations are referred to as naive, central memory, effector memory and effector memory RA+ (EMRA). The aim of this study was to identify potential lineage relationships between these subpopulations for both CD4+ and CD8+ T cells through microarray analysis. The genes found to distinguish between these subpopulations include many molecules with known functions in T cell differentiation, including CCR7, CD45RA, granzymes, L-selectin and TNF receptors. Several genes from the tetraspanin family of proteins were found to be differentially expressed at mRNA and protein level; suggesting a possible role for these genes in CD4+ and CD8+ T cell activation, migration and lysosomal function. -

Neural Development
31 October 2007 NEURAL DEVELOPMENT www.neuraldevelopment.com Analysis of Lrrn1 expression and its relationship to neuromeric boundaries during chick neural development Laura C Andreae et al. Neural Development 2007, 2:22 http://www.neuraldevelopment.com/content/2/1/22 Neural Development BioMed Central Research article Open Access Analysis of Lrrn1 expression and its relationship to neuromeric boundaries during chick neural development LauraCAndreae1,2, Daniela Peukert1, Andrew Lumsden1 and Jonathan D Gilthorpe*1 Address: 1MRC Centre for Developmental Neurobiology, King's College London, New Hunt's House, Guy's Campus, London, UK, SE1 1UL and 2Department of Neurophysiology, National Institute for Medical Research, The Ridgeway, Mill Hill, London, UK, NW7 1AA Email: Laura C Andreae - [email protected]; Daniela Peukert - [email protected]; Andrew Lumsden - [email protected]; Jonathan D Gilthorpe* - [email protected] * Corresponding author Published: 31 October 2007 Received: 26 March 2007 Accepted: 31 October 2007 Neural Development 2007, 2:22 doi:10.1186/1749-8104-2-22 This article is available from: http://www.neuraldevelopment.com/content/2/1/22 © 2007 Andreae et al.; licensee BioMed Central Ltd. This is an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: The Drosophila leucine-rich repeat proteins Tartan (TRN) and Capricious (CAPS) mediate cell affinity differences during compartition of the wing imaginal disc. This study aims to identify and characterize the expression of a chick orthologue of TRN/CAPS and examine its potential function in relation to compartment boundaries in the vertebrate central nervous system. -

Epigenetic Mechanisms Are Involved in the Oncogenic Properties of ZNF518B in Colorectal Cancer
Epigenetic mechanisms are involved in the oncogenic properties of ZNF518B in colorectal cancer Francisco Gimeno-Valiente, Ángela L. Riffo-Campos, Luis Torres, Noelia Tarazona, Valentina Gambardella, Andrés Cervantes, Gerardo López-Rodas, Luis Franco and Josefa Castillo SUPPLEMENTARY METHODS 1. Selection of genomic sequences for ChIP analysis To select the sequences for ChIP analysis in the five putative target genes, namely, PADI3, ZDHHC2, RGS4, EFNA5 and KAT2B, the genomic region corresponding to the gene was downloaded from Ensembl. Then, zoom was applied to see in detail the promoter, enhancers and regulatory sequences. The details for HCT116 cells were then recovered and the target sequences for factor binding examined. Obviously, there are not data for ZNF518B, but special attention was paid to the target sequences of other zinc-finger containing factors. Finally, the regions that may putatively bind ZNF518B were selected and primers defining amplicons spanning such sequences were searched out. Supplementary Figure S3 gives the location of the amplicons used in each gene. 2. Obtaining the raw data and generating the BAM files for in silico analysis of the effects of EHMT2 and EZH2 silencing The data of siEZH2 (SRR6384524), siG9a (SRR6384526) and siNon-target (SRR6384521) in HCT116 cell line, were downloaded from SRA (Bioproject PRJNA422822, https://www.ncbi. nlm.nih.gov/bioproject/), using SRA-tolkit (https://ncbi.github.io/sra-tools/). All data correspond to RNAseq single end. doBasics = TRUE doAll = FALSE $ fastq-dump -I --split-files SRR6384524 Data quality was checked using the software fastqc (https://www.bioinformatics.babraham. ac.uk /projects/fastqc/). The first low quality removing nucleotides were removed using FASTX- Toolkit (http://hannonlab.cshl.edu/fastxtoolkit/). -

Study Implicates Cell-Adhesion Proteins in Autism
Spectrum | Autism Research News https://www.spectrumnews.org NEWS Study implicates cell-adhesion proteins in autism BY VICTORIA STERN 18 MAY 2010 Sticky clues: Variations in leucine-rich repeat genes, which produce cell-adhesion proteins, could be important in autism. Variations in two genes needed to form connections between brain cells may be associated with autism spectrum disorder, according to a study published 25 March in Molecular Autism1. Some variants in the genes increase susceptibility to autism, whereas others protect children from developing the disorder. Several studies in people and mice have linked autism to proteins that build synapses, the junction between neurons. Defects in these proteins, including neurexin 1 and neuroligin-4, have been associated with a range of autistism-like behaviors, including impaired social interactions and problems with learning and memory. "This study fits nicely with the current literature, which says that genes in charge of synaptic function and development in the brain may be relevant for autism," says Hakon Hakonarson, director of the Center for Applied Genomics at The Children's Hospital of Philadelphia, who was not involved with the study. 1 / 4 Spectrum | Autism Research News https://www.spectrumnews.org The investigators focused on a group of genes called leucine-rich repeat (LRR) genes, which produce cell-adhesion proteins. These are molecules located on the neuron's surface that help one neuron bond to another, forming a network of connections between the cells. In the past few years, defects in these proteins have been linked to neurological disorders such as Parkinson's disease, epilepsy2 and autism3. -

BMC Medical Genetics Biomed Central
BMC Medical Genetics BioMed Central Research article Open Access Examination of NRCAM, LRRN3, KIAA0716, and LAMB1 as autism candidate genes Holli B Hutcheson1,2, Lana M Olson1, Yuki Bradford1, Susan E Folstein3, Susan L Santangelo4, James S Sutcliffe1 and Jonathan L Haines*1 Address: 1Center for Human Genetics Research and Department of Molecular Physiology and Biophysics, Vanderbilt University Medical Center, Nashville, TN, 37027, U.S.A, 2Laboratory of Genomic Diversity, National Cancer Institute, Frederick Cancer Research and Development Center, Frederick, MD, 21702, U.S.A, 3Department of Psychiatry, New England Medical Center/Tufts University School of Medicine, Boston, MA, 02111, U.S.A and 4Psychiatric & Neurodevelopmental Genetics Unit, Massachusetts General Hospital, Boston, MA, 02129, U.S.A Email: Holli B Hutcheson - [email protected]; Lana M Olson - [email protected]; Yuki Bradford - [email protected]; Susan E Folstein - [email protected]; Susan L Santangelo - [email protected]; James S Sutcliffe - [email protected]; Jonathan L Haines* - [email protected] * Corresponding author Published: 05 May 2004 Received: 20 November 2003 Accepted: 05 May 2004 BMC Medical Genetics 2004, 5:12 This article is available from: http://www.biomedcentral.com/1471-2350/5/12 © 2004 Hutcheson et al; licensee BioMed Central Ltd. This is an Open Access article: verbatim copying and redistribution of this article are permitted in all media for any purpose, provided this notice is preserved along with the article's original URL. Abstract Background: A substantial body of research supports a genetic involvement in autism. Furthermore, results from various genomic screens implicate a region on chromosome 7q31 as harboring an autism susceptibility variant. -

Gene Expression in Blood Reflects Smoking Exposure Among Cancer
www.nature.com/scientificreports OPEN Gene expression in blood refects smoking exposure among cancer‑free women in the Norwegian Women and Cancer (NOWAC) postgenome cohort Nikita Baiju1*, Torkjel M. Sandanger1, Pål Sætrom2,3,4,5 & Therese H. Nøst1,5 Active smoking has been linked to modulated gene expression in blood. However, there is a need for a more thorough understanding of how quantitative measures of smoking exposure relate to diferentially expressed genes (DEGs) in whole‑blood among ever smokers. This study analysed microarray‑based gene expression profles from whole‑blood samples according to smoking status and quantitative measures of smoking exposure among cancer‑free women (n = 1708) in the Norwegian Women and Cancer postgenome cohort. When compared with never smokers and former smokers, current smokers had 911 and 1082 DEGs, respectively and their biological functions could indicate systemic impacts of smoking. LRRN3 was associated with smoking status with the lowest FDR‑adjusted p‑value. When never smokers and all former smokers were compared, no DEGs were observed, but LRRN3 was diferentially expressed when never smokers were compared with former smokers who quit smoking ≤ 10 years ago. Further, LRRN3 was positively associated with smoking intensity, pack‑years, and comprehensive smoking index score among current smokers; and negatively associated with time since cessation among former smokers. Consequently, LRRN3 expression in whole‑blood is a molecular signal of smoking exposure that could supplant self‑reported smoking data in further research targeting blood‑based markers related to the health efects of smoking. Tobacco smoking is one of the major threats to public health, and it is currently responsible for more than 8 million deaths worldwide each year 1. -

(B6;129.Cg-Gt(ROSA)26Sor Tm20(CAG-Ctgf-GFP)Jsd) Were Crossed with Female Foxd1cre/+ Heterozygote Mice 1, and Experimental Mice Were Selected As Foxd1cre/+; Rs26cig/+
Supplemental Information SI Methods Animal studies Heterozygote mice (B6;129.Cg-Gt(ROSA)26Sor tm20(CAG-Ctgf-GFP)Jsd) were crossed with female Foxd1Cre/+ heterozygote mice 1, and experimental mice were selected as Foxd1Cre/+; Rs26CIG/+. In some studies Coll-GFPTg or TCF/Lef:H2B-GFPTg mice or Foxd1Cre/+; Rs26tdTomatoR/+ mice were used as described 2; 3. Left kidneys were subjected to ureteral obstruction using a posterior surgical approach as described 2. In some experiments recombinant mouse DKK1 (0.5mg/kg) or an equal volume of vehicle was administered by daily IP injection. In the in vivo ASO experiment, either specific Lrp6 (TACCTCAATGCGATTT) or scrambled negative control ASO (AACACGTCTATACGC) (30mg/kg) (Exiqon, LNA gapmers) was administered by IP injection on d-1, d1, d4, and d7. In other experiments anti-CTGF domain-IV antibodies (5mg/kg) or control IgG were administered d-1, d1 and d6. All animal experiments were performed under approved IACUC protocols held at the University of Washington and Biogen. Recombinant protein and antibody generation and characterization Human CTGF domain I (sequence Met1 CPDEPAPRCPAGVSLVLDGCGCCRVCAKQLGELCTERDPCDPHKGLFC), domain I+II (sequence Met1CPDEPAPRCPAGVSLVLDGCGCCRVCAKQLGELCTERDPCDPHKGLFCCIFGGT VYRSGESFQSSCKYQCTCLDGAVGCMPLCSMDVRLPSPDCPFPRRVKLPGKCCEE) were cloned and expressed in 293 cells, and purified by Chelating SFF(Ni) Column, tested for single band by SEC and PAGE, and tested for absence of contamination. Domain-IV (sequence GKKCIRTPKISKPIKFELSGCTSMKTYRAKFCGVCTDGRCCTPHRTTTLPVEFKCPDGE VMKKNMMFIKTCACHYNCPGDNDIFESLYYRKMY) was purchased from Peprotech. Mouse or human DKK1 was generated from the coding sequence with some modifications and a tag. Secreted protein was harvested from 293 cells, and purified by nickel column, and tested for activity in a supertopflash (STF) assay 4. DKK1 showed EC50 of 0.69nM for WNT3a-induced WNT signaling in STF cells.