Neural Development
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Table 2. Functional Classification of Genes Differentially Regulated After HOXB4 Inactivation in HSC/Hpcs
Table 2. Functional classification of genes differentially regulated after HOXB4 inactivation in HSC/HPCs Symbol Gene description Fold-change (mean ± SD) Signal transduction Adam8 A disintegrin and metalloprotease domain 8 1.91 ± 0.51 Arl4 ADP-ribosylation factor-like 4 - 1.80 ± 0.40 Dusp6 Dual specificity phosphatase 6 (Mkp3) - 2.30 ± 0.46 Ksr1 Kinase suppressor of ras 1 1.92 ± 0.42 Lyst Lysosomal trafficking regulator 1.89 ± 0.34 Mapk1ip1 Mitogen activated protein kinase 1 interacting protein 1 1.84 ± 0.22 Narf* Nuclear prelamin A recognition factor 2.12 ± 0.04 Plekha2 Pleckstrin homology domain-containing. family A. (phosphoinosite 2.15 ± 0.22 binding specific) member 2 Ptp4a2 Protein tyrosine phosphatase 4a2 - 2.04 ± 0.94 Rasa2* RAS p21 activator protein 2 - 2.80 ± 0.13 Rassf4 RAS association (RalGDS/AF-6) domain family 4 3.44 ± 2.56 Rgs18 Regulator of G-protein signaling - 1.93 ± 0.57 Rrad Ras-related associated with diabetes 1.81 ± 0.73 Sh3kbp1 SH3 domain kinase bindings protein 1 - 2.19 ± 0.53 Senp2 SUMO/sentrin specific protease 2 - 1.97 ± 0.49 Socs2 Suppressor of cytokine signaling 2 - 2.82 ± 0.85 Socs5 Suppressor of cytokine signaling 5 2.13 ± 0.08 Socs6 Suppressor of cytokine signaling 6 - 2.18 ± 0.38 Spry1 Sprouty 1 - 2.69 ± 0.19 Sos1 Son of sevenless homolog 1 (Drosophila) 2.16 ± 0.71 Ywhag 3-monooxygenase/tryptophan 5- monooxygenase activation protein. - 2.37 ± 1.42 gamma polypeptide Zfyve21 Zinc finger. FYVE domain containing 21 1.93 ± 0.57 Ligands and receptors Bambi BMP and activin membrane-bound inhibitor - 2.94 ± 0.62 -

Anti-Lunatic Fringe Antibody (ARG40136)
Product datasheet [email protected] ARG40136 Package: 100 μl anti-Lunatic Fringe antibody Store at: -20°C Summary Product Description Rabbit Polyclonal antibody recognizes Lunatic Fringe Tested Reactivity Hu Tested Application FACS, IHC-P, WB Host Rabbit Clonality Polyclonal Isotype IgG Target Name Lunatic Fringe Antigen Species Human Immunogen KLH-conjugated synthetic peptide corresponding to aa. 86-114 of Human Lunatic Fringe. Conjugation Un-conjugated Alternate Names O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase; EC 2.4.1.222; SCDO3; Beta-1,3-N- acetylglucosaminyltransferase lunatic fringe Application Instructions Application table Application Dilution FACS 1:10 - 1:50 IHC-P 1:50 - 1:100 WB 1:1000 Application Note * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. Positive Control K562 Calculated Mw 42 kDa Properties Form Liquid Purification Purification with Protein A and immunogen peptide. Buffer PBS and 0.09% (W/V) Sodium azide. Preservative 0.09% (W/V) Sodium azide. Storage instruction For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C or below. Storage in frost free freezers is not recommended. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. www.arigobio.com 1/3 Note For laboratory research only, not for drug, diagnostic or other use. Bioinformation Gene Symbol LFNG Gene Full Name LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase Background This gene is a member of the fringe gene family which also includes radical and manic fringe genes. -

Genomic Correlates of Relationship QTL Involved in Fore- Versus Hind Limb Divergence in Mice
Loyola University Chicago Loyola eCommons Biology: Faculty Publications and Other Works Faculty Publications 2013 Genomic Correlates of Relationship QTL Involved in Fore- Versus Hind Limb Divergence in Mice Mihaela Palicev Gunter P. Wagner James P. Noonan Benedikt Hallgrimsson James M. Cheverud Loyola University Chicago, [email protected] Follow this and additional works at: https://ecommons.luc.edu/biology_facpubs Part of the Biology Commons Recommended Citation Palicev, M, GP Wagner, JP Noonan, B Hallgrimsson, and JM Cheverud. "Genomic Correlates of Relationship QTL Involved in Fore- Versus Hind Limb Divergence in Mice." Genome Biology and Evolution 5(10), 2013. This Article is brought to you for free and open access by the Faculty Publications at Loyola eCommons. It has been accepted for inclusion in Biology: Faculty Publications and Other Works by an authorized administrator of Loyola eCommons. For more information, please contact [email protected]. This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 3.0 License. © Palicev et al., 2013. GBE Genomic Correlates of Relationship QTL Involved in Fore- versus Hind Limb Divergence in Mice Mihaela Pavlicev1,2,*, Gu¨ nter P. Wagner3, James P. Noonan4, Benedikt Hallgrı´msson5,and James M. Cheverud6 1Konrad Lorenz Institute for Evolution and Cognition Research, Altenberg, Austria 2Department of Pediatrics, Cincinnati Children‘s Hospital Medical Center, Cincinnati, Ohio 3Yale Systems Biology Institute and Department of Ecology and Evolutionary Biology, Yale University 4Department of Genetics, Yale University School of Medicine 5Department of Cell Biology and Anatomy, The McCaig Institute for Bone and Joint Health and the Alberta Children’s Hospital Research Institute for Child and Maternal Health, University of Calgary, Calgary, Canada 6Department of Anatomy and Neurobiology, Washington University *Corresponding author: E-mail: [email protected]. -

Segmentation in Vertebrates: Clock and Gradient Finally Joined
Downloaded from genesdev.cshlp.org on September 24, 2021 - Published by Cold Spring Harbor Laboratory Press REVIEW Segmentation in vertebrates: clock and gradient finally joined Alexander Aulehla1 and Bernhard G. Herrmann2 Max-Planck-Institute for Molecular Genetics, Department of Developmental Genetics, 14195 Berlin, Germany The vertebral column is derived from somites formed by terior (A–P) axis. Somite formation takes place periodi- segmentation of presomitic mesoderm, a fundamental cally in a fixed anterior-to-posterior sequence. process of vertebrate embryogenesis. Models on the In the chick embryo, a new somite is formed approxi- mechanism controlling this process date back some mately every 90 min, whereas in the mouse embryo, the three to four decades. Access to understanding the mo- periodicity varies, dependent on the axial position (Tam lecular control of somitogenesis has been gained only 1981). Classical embryology experiments revealed that recently by the discovery of molecular oscillators (seg- periodicity and directionality of somite formation are mentation clock) and gradients of signaling molecules, controlled by an intrinsic program set off in the cells as as predicted by early models. The Notch signaling path- they are recruited into the psm. For instance, when the way is linked to the oscillator and plays a decisive role in psm is inverted rostro–caudally, somite formation in the inter- and intrasomitic boundary formation. An Fgf8 sig- inverted region proceeds from caudal to rostral, main- naling gradient is involved in somite size control. And taining the original direction (Christ et al. 1974). More- the (canonical) Wnt signaling pathway, driven by Wnt3a, over, neither the transversal bisection nor the isolation appears to integrate clock and gradient in a global of the psm from all surrounding tissues stops the seg- mechanism controlling the segmentation process. -

Whole-Genome Microarray Detects Deletions and Loss of Heterozygosity of Chromosome 3 Occurring Exclusively in Metastasizing Uveal Melanoma
Anatomy and Pathology Whole-Genome Microarray Detects Deletions and Loss of Heterozygosity of Chromosome 3 Occurring Exclusively in Metastasizing Uveal Melanoma Sarah L. Lake,1 Sarah E. Coupland,1 Azzam F. G. Taktak,2 and Bertil E. Damato3 PURPOSE. To detect deletions and loss of heterozygosity of disease is fatal in 92% of patients within 2 years of diagnosis. chromosome 3 in a rare subset of fatal, disomy 3 uveal mela- Clinical and histopathologic risk factors for UM metastasis noma (UM), undetectable by fluorescence in situ hybridization include large basal tumor diameter (LBD), ciliary body involve- (FISH). ment, epithelioid cytomorphology, extracellular matrix peri- ϩ ETHODS odic acid-Schiff-positive (PAS ) loops, and high mitotic M . Multiplex ligation-dependent probe amplification 3,4 5 (MLPA) with the P027 UM assay was performed on formalin- count. Prescher et al. showed that a nonrandom genetic fixed, paraffin-embedded (FFPE) whole tumor sections from 19 change, monosomy 3, correlates strongly with metastatic death, and the correlation has since been confirmed by several disomy 3 metastasizing UMs. Whole-genome microarray analy- 3,6–10 ses using a single-nucleotide polymorphism microarray (aSNP) groups. Consequently, fluorescence in situ hybridization were performed on frozen tissue samples from four fatal dis- (FISH) detection of chromosome 3 using a centromeric probe omy 3 metastasizing UMs and three disomy 3 tumors with Ͼ5 became routine practice for UM prognostication; however, 5% years’ metastasis-free survival. to 20% of disomy 3 UM patients unexpectedly develop metas- tases.11 Attempts have therefore been made to identify the RESULTS. Two metastasizing UMs that had been classified as minimal region(s) of deletion on chromosome 3.12–15 Despite disomy 3 by FISH analysis of a small tumor sample were found these studies, little progress has been made in defining the key on MLPA analysis to show monosomy 3. -

A Conserved Gene Family Encodes Transmembrane Proteins with Fibronectin, Immunoglobulin and Leucine-Rich Repeat Domains (FIGLER) Delicia L
A conserved gene family encodes transmembrane proteins with fibronectin, immunoglobulin and leucine-rich repeat domains (FIGLER) Delicia L. Munfus, University of Alabama at Birmingham Christopher L. Haga, University of Alabama at Birmingham Peter D. Burrows, University of Alabama at Birmingham Max Cooper, Emory University Journal Title: BMC Biology Volume: Volume 5, Number 1 Publisher: BioMed Central | 2007-09-13, Pages 36-36 Type of Work: Article | Final Publisher PDF Publisher DOI: 10.1186/1741-7007-5-36 Permanent URL: https://pid.emory.edu/ark:/25593/rqdhh Final published version: http://dx.doi.org/10.1186/1741-7007-5-36 Copyright information: © 2007 Munfus et al; licensee BioMed Central Ltd. This is an Open Access work distributed under the terms of the Creative Commons Attribution 2.0 Generic License (http://creativecommons.org/licenses/by/2.0/). Accessed October 6, 2021 6:19 AM EDT BMC Biology BioMed Central Research article Open Access A conserved gene family encodes transmembrane proteins with fibronectin, immunoglobulin and leucine-rich repeat domains (FIGLER) Delicia L Munfus†1,2, Christopher L Haga†1,2, Peter D Burrows1,2,3 and Max D Cooper*1,2,4,5 Address: 1Division of Developmental and Clinical Immunology, University of Alabama at Birmingham, Birmingham, AL 35294-3300, USA, 2Department of Microbiology, University of Alabama at Birmingham, Birmingham, AL 35294-3300, USA, 3Department of Genetics, University of Alabama at Birmingham, Birmingham, AL 35294-3300,, 4Department of Medicine, University of Alabama at Birmingham, Birmingham, -

Supplemental Table 1. Complete Gene Lists and GO Terms from Figure 3C
Supplemental Table 1. Complete gene lists and GO terms from Figure 3C. Path 1 Genes: RP11-34P13.15, RP4-758J18.10, VWA1, CHD5, AZIN2, FOXO6, RP11-403I13.8, ARHGAP30, RGS4, LRRN2, RASSF5, SERTAD4, GJC2, RHOU, REEP1, FOXI3, SH3RF3, COL4A4, ZDHHC23, FGFR3, PPP2R2C, CTD-2031P19.4, RNF182, GRM4, PRR15, DGKI, CHMP4C, CALB1, SPAG1, KLF4, ENG, RET, GDF10, ADAMTS14, SPOCK2, MBL1P, ADAM8, LRP4-AS1, CARNS1, DGAT2, CRYAB, AP000783.1, OPCML, PLEKHG6, GDF3, EMP1, RASSF9, FAM101A, STON2, GREM1, ACTC1, CORO2B, FURIN, WFIKKN1, BAIAP3, TMC5, HS3ST4, ZFHX3, NLRP1, RASD1, CACNG4, EMILIN2, L3MBTL4, KLHL14, HMSD, RP11-849I19.1, SALL3, GADD45B, KANK3, CTC- 526N19.1, ZNF888, MMP9, BMP7, PIK3IP1, MCHR1, SYTL5, CAMK2N1, PINK1, ID3, PTPRU, MANEAL, MCOLN3, LRRC8C, NTNG1, KCNC4, RP11, 430C7.5, C1orf95, ID2-AS1, ID2, GDF7, KCNG3, RGPD8, PSD4, CCDC74B, BMPR2, KAT2B, LINC00693, ZNF654, FILIP1L, SH3TC1, CPEB2, NPFFR2, TRPC3, RP11-752L20.3, FAM198B, TLL1, CDH9, PDZD2, CHSY3, GALNT10, FOXQ1, ATXN1, ID4, COL11A2, CNR1, GTF2IP4, FZD1, PAX5, RP11-35N6.1, UNC5B, NKX1-2, FAM196A, EBF3, PRRG4, LRP4, SYT7, PLBD1, GRASP, ALX1, HIP1R, LPAR6, SLITRK6, C16orf89, RP11-491F9.1, MMP2, B3GNT9, NXPH3, TNRC6C-AS1, LDLRAD4, NOL4, SMAD7, HCN2, PDE4A, KANK2, SAMD1, EXOC3L2, IL11, EMILIN3, KCNB1, DOK5, EEF1A2, A4GALT, ADGRG2, ELF4, ABCD1 Term Count % PValue Genes regulation of pathway-restricted GDF3, SMAD7, GDF7, BMPR2, GDF10, GREM1, BMP7, LDLRAD4, SMAD protein phosphorylation 9 6.34 1.31E-08 ENG pathway-restricted SMAD protein GDF3, SMAD7, GDF7, BMPR2, GDF10, GREM1, BMP7, LDLRAD4, phosphorylation -

Genetic Drivers of Pancreatic Islet Function
| INVESTIGATION Genetic Drivers of Pancreatic Islet Function Mark P. Keller,*,1 Daniel M. Gatti,†,1 Kathryn L. Schueler,* Mary E. Rabaglia,* Donnie S. Stapleton,* Petr Simecek,† Matthew Vincent,† Sadie Allen,‡ Aimee Teo Broman,§ Rhonda Bacher,§ Christina Kendziorski,§ Karl W. Broman,§ Brian S. Yandell,** Gary A. Churchill,†,2 and Alan D. Attie*,2 *Department of Biochemistry, §Department of Biostatistics and Medical Informatics, and **Department of Horticulture, University of Wisconsin–Madison, Wisconsin 53706-1544, †The Jackson Laboratory, Bar Harbor, Maine 06409, and ‡Maine School of Science and Mathematics, Limestone, Maine 06409, ORCID IDs: 0000-0002-7405-5552 (M.P.K.); 0000-0002-4914-6671 (K.W.B.); 0000-0001-9190-9284 (G.A.C.); 0000-0002-0568-2261 (A.D.A.) ABSTRACT The majority of gene loci that have been associated with type 2 diabetes play a role in pancreatic islet function. To evaluate the role of islet gene expression in the etiology of diabetes, we sensitized a genetically diverse mouse population with a Western diet high in fat (45% kcal) and sucrose (34%) and carried out genome-wide association mapping of diabetes-related phenotypes. We quantified mRNA abundance in the islets and identified 18,820 expression QTL. We applied mediation analysis to identify candidate causal driver genes at loci that affect the abundance of numerous transcripts. These include two genes previously associated with monogenic diabetes (PDX1 and HNF4A), as well as three genes with nominal association with diabetes-related traits in humans (FAM83E, IL6ST, and SAT2). We grouped transcripts into gene modules and mapped regulatory loci for modules enriched with transcripts specific for a-cells, and another specific for d-cells. -

Epigenetic Alterations of Chromosome 3 Revealed by Noti-Microarrays in Clear Cell Renal Cell Carcinoma
Hindawi Publishing Corporation BioMed Research International Volume 2014, Article ID 735292, 9 pages http://dx.doi.org/10.1155/2014/735292 Research Article Epigenetic Alterations of Chromosome 3 Revealed by NotI-Microarrays in Clear Cell Renal Cell Carcinoma Alexey A. Dmitriev,1,2 Evgeniya E. Rudenko,3 Anna V. Kudryavtseva,1,2 George S. Krasnov,1,4 Vasily V. Gordiyuk,3 Nataliya V. Melnikova,1 Eduard O. Stakhovsky,5 Oleksii A. Kononenko,5 Larissa S. Pavlova,6 Tatiana T. Kondratieva,6 Boris Y. Alekseev,2 Eleonora A. Braga,7,8 Vera N. Senchenko,1 and Vladimir I. Kashuba3,9 1 Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, Moscow 119991, Russia 2 P.A. Herzen Moscow Oncology Research Institute, Ministry of Healthcare of the Russian Federation, Moscow 125284, Russia 3 Institute of Molecular Biology and Genetics, Ukrainian Academy of Sciences, Kiev 03680, Ukraine 4 Mechnikov Research Institute for Vaccines and Sera, Russian Academy of Medical Sciences, Moscow 105064, Russia 5 National Cancer Institute, Kiev 03022, Ukraine 6 N.N. Blokhin Russian Cancer Research Center, Russian Academy of Medical Sciences, Moscow 115478, Russia 7 Institute of General Pathology and Pathophysiology, Russian Academy of Medical Sciences, Moscow 125315, Russia 8 Research Center of Medical Genetics, Russian Academy of Medical Sciences, Moscow 115478, Russia 9 DepartmentofMicrobiology,TumorandCellBiology,KarolinskaInstitute,17177Stockholm,Sweden Correspondence should be addressed to Alexey A. Dmitriev; alex [email protected] Received 19 February 2014; Revised 10 April 2014; Accepted 17 April 2014; Published 22 May 2014 Academic Editor: Carole Sourbier Copyright © 2014 Alexey A. Dmitriev et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. -

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -

Single-Cell RNA-Seq of Dopaminergic Neurons Informs Candidate Gene Selection for Sporadic
bioRxiv preprint doi: https://doi.org/10.1101/148049; this version posted August 31, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Single-cell RNA-seq of dopaminergic neurons informs candidate gene selection for sporadic 2 Parkinson's disease 3 4 Paul W. Hook1, Sarah A. McClymont1, Gabrielle H. Cannon1, William D. Law1, A. Jennifer 5 Morton2, Loyal A. Goff1,3*, Andrew S. McCallion1,4,5* 6 7 1McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of 8 Medicine, Baltimore, Maryland, United States of America 9 2Department of Physiology Development and Neuroscience, University of Cambridge, 10 Cambridge, United Kingdom 11 3Department of Neuroscience, Johns Hopkins University School of Medicine, Baltimore, 12 Maryland, United States of America 13 4Department of Comparative and Molecular Pathobiology, Johns Hopkins University School of 14 Medicine, Baltimore, Maryland, United States of America 15 5Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, 16 United States of America 17 *, To whom correspondence should be addressed: [email protected] and [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/148049; this version posted August 31, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. -
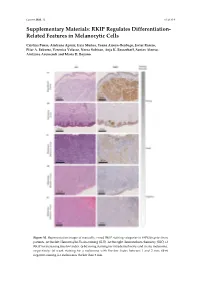
RKIP Regulates Differentiation- Related Features in Melanocytic Cells
Cancers 2020. 12 S1 of S14 Supplementary Materials: RKIP Regulates Differentiation- Related Features in Melanocytic Cells Cristina Penas, Aintzane Apraiz, Iraia Muñoa, Yoana Arroyo-Berdugo, Javier Rasero, Pilar A. Ezkurra, Veronica Velasco, Nerea Subiran, Anja K. Bosserhoff, Santos Alonso, Aintzane Asumendi and Maria D. Boyano Figure S1. Representative images of manually scored RKIP staining categories in FFPE biopsies from patients. At the feft: Hematoxylin-Eosin staining (H-E). At the right: Immunohistochemistry (IHC) of RKIP for increasing Breslow Index. (a-b) Strong staining for intradermal nevus and in situ melanoma, respectively; (c) weak staining for a melanoma with Breslow Index between 1 and 2 mm; (d-e) negative staining for melanomas thicker than 2 mm. Cancers 2020. 12 S2 of S14 Figure S2. RNA Sequencing data analysis. Each row of the figure represents an over-represented gene signature generated by analyzing the up-regulated and down-regulated genes from a differential expression analysis of RKIP Knockdown melanocytes HEMn-LP. Results from the EnrichR Website with a p-value > 0.05. Figure S3. RNA Sequencing data quality. (a) Read counts normalization per sample; (b) Spearman correlation coefficient between replicates; (c) Principal component analysis for replicates. Cancers 2020. 12 S3 of S14 Figure S4. Details of Western Blot from Figure 2 related with RKIP expression in several cell lines. (a) Blots for RKIP and Tubulin in primary and metastatic melanoma and in three primary melanocytes HEMn-DP (dark pigmented), HEMn-LP (light pigmented) and HEM2710. On the right, original blot for each analyzed protein. Highlighted with a red square the information included in the Figure 3 (horizontal rotation).