Mechanisms of Resistance to Β-Lactam Antibiotics in Streptomycetes Wedad
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Synthesis and Biological Evaluation of Trisindolyl-Cycloalkanes and Bis- Indolyl Naphthalene Small Molecules As Potent Antibacterial and Antifungal Agents
Synthesis and Biological Evaluation of Trisindolyl-Cycloalkanes and Bis- Indolyl Naphthalene Small Molecules as Potent Antibacterial and Antifungal Agents Dissertation Zur Erlangung des akademischen Grades doctor rerum naturalium (Dr. rer. nat.) Vorgelegt der Naturwissenschaftlichen Fakultät I Institut für Pharmazie Fachbereich für Pharmazeutische Chemie der Martin-Luther-Universität Halle-Wittenberg von Kaveh Yasrebi Geboren am 09.14.1987 in Teheran/Iran (Islamische Republik) Gutachter: 1. Prof. Dr. Andreas Hilgeroth (Martin-Luther-Universität Halle-Wittenberg, Germany) 2. Prof. Dr. Sibel Süzen (Ankara Üniversitesi, Turkey) 3. Prof. Dr. Michael Lalk (Ernst-Moritz-Arndt-Universität Greifswald, Germany) Halle (Saale), den 21. Juli 2020 Selbstständigkeitserklärung Hiermit erkläre ich gemäß § 5 (2) b der Promotionsordnung der Naturwissenschaftlichen Fakultät I – Institut für Pharmazie der Martin-Luther-Universität Halle-Wittenberg, dass ich die vorliegende Arbeit selbstständig und ohne Benutzung anderer als der angegebenen Hilfsmittel und Quellen angefertigt habe. Alle Stellen, die wörtlich oder sinngemäß aus Veröffentlichungen entnommen sind, habe ich als solche kenntlich gemacht. Ich erkläre ferner, dass diese Arbeit in gleicher oder ähnlicher Form bisher keiner anderen Prüfbehörde zur Erlangung des Doktorgrades vorgelegt wurde. Halle (Saale), den 21. Juli 2020 Kaveh Yasrebi Acknowledgement This study was carried out from June 2015 to July 2017 in the Research Group of Drug Development and Analysis led by Prof. Dr. Andreas Hilgeroth at the Institute of Pharmacy, Martin-Luther-Universität Halle-Wittenberg. I would like to thank all the people for their participation who supported my work in this way and helped me obtain good results. First of all, I would like to express my gratitude to Prof. Dr. Andreas Hilgeroth for providing me with opportunity to carry out my Ph.D. -

Novel Antimicrobial Agents Inhibiting Lipid II Incorporation Into Peptidoglycan Essay MBB
27 -7-2019 Novel antimicrobial agents inhibiting lipid II incorporation into peptidoglycan Essay MBB Mark Nijland S3265978 Supervisor: Prof. Dr. Dirk-Jan Scheffers Molecular Microbiology University of Groningen Content Abstract..............................................................................................................................................2 1.0 Peptidoglycan biosynthesis of bacteria ........................................................................................3 2.0 Novel antimicrobial agents ...........................................................................................................4 2.1 Teixobactin ...............................................................................................................................4 2.2 tridecaptin A1............................................................................................................................7 2.3 Malacidins ................................................................................................................................8 2.4 Humimycins ..............................................................................................................................9 2.5 LysM ........................................................................................................................................ 10 3.0 Concluding remarks .................................................................................................................... 11 4.0 references ................................................................................................................................. -
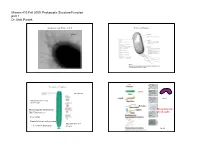
Parsek Micro410 Lecture
Microm 410 Fall 2009: Prokaryotic Structure/Function part 1 Dr. Matt Parsek Organization of the Prokaryotic Cell Prokaryotic Structures fimbriae Size Range of Prokaryotes bacillus See Table 4.1 (rigid) vibrio Nanobacteria 0.05‐0.2 µm (0.14‐0.2 µm) (flexible) Thiomargarita namibiensis Mycoplasma are 700‐750 µm (Fig. 4.2) pleomorphic green alga Nanochlorum eukaryotum Mycoplasma 0.1‐ ~1-2 µm in diameter 0.3 µm Fig. 4.1 Microm 410 Fall 2009: Prokaryotic Structure/Function part 1 Dr. Matt Parsek Staining cells for Microscopic observation Cell Arrangements Motility- ~80% of prokaryotes are motile streptococcus Staining properties: Gram Stain staphylococcus Fig. 2.3 Gram Stain (1884) (Bacteria) Gram-negative mixed culture Gram-positive Fig. 2.3 and 2.4 Microm 410 Fall 2009: Prokaryotic Structure/Function part 1 Dr. Matt Parsek Functions of the cytoplasmic membrane The phospholipid bi‐layer Fig. 4.9 Fig. 4.4 What is the structure of bacterial phospholipids? Other components of the cytoplasmic membrane Figs. 4.5‐4.6 Microm 410 Fall 2009: Prokaryotic Structure/Function part 1 Dr. Matt Parsek Archaeal membranes can be a lipid monolayer Archaeal phospholipids have an ether linkage Fig. 4.7 Fig. 4.8 Importance of Cell Wall Schematic diagram cell wall • Provides rigidity to cell allowing cell to withstand the large osmotic/ionic Fig. 4.16 changes a bacterium may experience in its environment, and turgor pressure of cytoplasm (conc. of solutes in cytoplasm). Cell lysis • May have a role in shape determination. • Provides a barrier against certain toxic chemical and biological agents. • Site of action of some of the most commonly used antibiotics used to treat bacterial infections (penicillin family). -
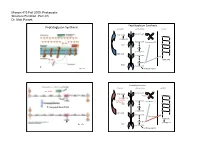
Parsek Lecture #3
Microm 410 Fall 2009: Prokaryotic Structure/Function: Part 2/3 Dr. Matt Parsek Peptidoglycan Synthesis Peptidoglycan Synthesis cytoplasm cell membrane cell wall Bactoprenol-P Pi UDP-NAM M G pentapeptide G M Bactoprenol Bactoprenol-P-P P M UMP G P NAM G M pentapeptide M G UDP-NAG Bactoprenol G P NAM‐NAG P NAM-NAG UMP pentapeptide Fig. 6.7a Interbridge peptide Peptidoglycan Synthesis Cross-linking of Peptidoglycan Strands cytoplasm cell membrane cell wall autolysins Bactoprenol-P Pi UDP-NAM Bacitracin M G pentapeptide G D-cycloserine Bactoprenol M (Oxamycin) Bactoprenol-P-P P M UMP G P Transpeptidase (FtsI) NAM G M pentapeptide M G UDP-NAG Bactoprenol G Vancomycin P NAM‐NAG P NAM-NAG pentapeptide UMP Fig. 6.7b pentapeptide Interbridge peptide Microm 410 Fall 2009: Prokaryotic Structure/Function: Part 2/3 Dr. Matt Parsek Cross-linking of Peptidoglycan Strands Antibiotic Resistance autolysins • Inactivate antibiotic β-lactamase (penicillinase) Clavulanic acid β-lactams Augmentin and Trimentin (combination of clavulanic acid and transpeptidase amoxicillin or ampicillin respectively) penicillins and cephalosporins lysozyme • Change chemistry of target site • Limit access of the antibiotic to target site Fig. 6.5 Cell Shape Determination • Modifications made to Peptidoglycan: ‐ lysozyme: Protoplasts/spheroplasts ‐ autolysins Bacillus subtilis ‐ endopeptidase Heliobacter pylori • Protein(s) may play a major role ‐ MreB protein Caulobacter crescentus ‐ MreB has homology to actin, a component of the cytoskeleton of eukaryotes. Shape determining protein‐ crescentin Fig. 6.4 Microm 410 Fall 2009: Prokaryotic Structure/Function: Part 2/3 Dr. Matt Parsek Cell Wall Gram-positive Bacteria intermediate filaments in the bacteria Caulobacter crescentus glycerol similar predicted structures of crescentin and intermediate filaments Fig. -

B-Lactams: Chemical Structure, Mode of Action and Mechanisms of Resistance
b-Lactams: chemical structure, mode of action and mechanisms of resistance Ru´ben Fernandes, Paula Amador and Cristina Prudeˆncio This synopsis summarizes the key chemical and bacteriological characteristics of b-lactams, penicillins, cephalosporins, carbanpenems, monobactams and others. Particular notice is given to first-generation to fifth-generation cephalosporins. This review also summarizes the main resistance mechanism to antibiotics, focusing particular attention to those conferring resistance to broad-spectrum cephalosporins by means of production of emerging cephalosporinases (extended-spectrum b-lactamases and AmpC b-lactamases), target alteration (penicillin-binding proteins from methicillin-resistant Staphylococcus aureus) and membrane transporters that pump b-lactams out of the bacterial cell. Keywords: b-lactams, chemical structure, mechanisms of resistance, mode of action Historical perspective Alexander Fleming first noticed the antibacterial nature of penicillin in 1928. When working with Antimicrobials must be understood as any kind of agent another bacteriological problem, Fleming observed with inhibitory or killing properties to a microorganism. a contaminated culture of Staphylococcus aureus with Antibiotic is a more restrictive term, which implies the the mold Penicillium notatum. Fleming remarkably saw natural source of the antimicrobial agent. Similarly, under- the potential of this unfortunate event. He dis- lying the term chemotherapeutic is the artificial origin of continued the work that he was dealing with and was an antimicrobial agent by chemical synthesis [1]. Initially, able to describe the compound around the mold antibiotics were considered as small molecular weight and isolates it. He named it penicillin and published organic molecules or metabolites used in response of his findings along with some applications of penicillin some microorganisms against others that inhabit the same [4]. -

WO 2015/028850 Al 5 March 2015 (05.03.2015) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2015/028850 Al 5 March 2015 (05.03.2015) P O P C T (51) International Patent Classification: AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, C07D 519/00 (2006.01) A61P 39/00 (2006.01) BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, C07D 487/04 (2006.01) A61P 35/00 (2006.01) DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, A61K 31/5517 (2006.01) A61P 37/00 (2006.01) HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KN, KP, KR, A61K 47/48 (2006.01) KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, (21) International Application Number: OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, PCT/IB2013/058229 SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, (22) International Filing Date: TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, 2 September 2013 (02.09.2013) ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (26) Publication Language: English GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, SZ, TZ, (71) Applicant: HANGZHOU DAC BIOTECH CO., LTD UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, [US/CN]; Room B2001-B2019, Building 2, No 452 Sixth TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, Street, Hangzhou Economy Development Area, Hangzhou EE, ES, FI, FR, GB, GR, HR, HU, IE, IS, IT, LT, LU, LV, City, Zhejiang 310018 (CN). -

View Details
INDEX CHAPTER NUMBER CHAPTER NAME PAGE Extraction of Fungal Chitosan and its Chapter-1 1-17 Advanced Application Isolation and Separation of Phenolics Chapter-2 using HPLC Tool: A Consolidate Survey 18-48 from the Plant System Advances in Microbial Genomics in Chapter-3 49-80 the Post-Genomics Era Advances in Biotechnology in the Chapter-4 81-94 Post Genomics era Plant Growth Promotion by Endophytic Chapter-5 Actinobacteria Associated with 95-107 Medicinal Plants Viability of Probiotics in Dairy Products: A Chapter-6 Review Focusing on Yogurt, Ice 108-132 Cream, and Cheese Published in: Dec 2018 Online Edition available at: http://openaccessebooks.com/ Reprints request: [email protected] Copyright: @ Corresponding Author Advances in Biotechnology Chapter 1 Extraction of Fungal Chitosan and its Advanced Application Sahira Nsayef Muslim1; Israa MS AL-Kadmy1*; Alaa Naseer Mohammed Ali1; Ahmed Sahi Dwaish2; Saba Saadoon Khazaal1; Sraa Nsayef Muslim3; Sarah Naji Aziz1 1Branch of Biotechnology, Department of Biology, College of Science, AL-Mustansiryiah University, Baghdad-Iraq 2Branch of Fungi and Plant Science, Department of Biology, College of Science, AL-Mustansiryiah University, Baghdad-Iraq 3Department of Geophysics, College of remote sensing and geophysics, AL-Karkh University for sci- ence, Baghdad-Iraq *Correspondense to: Israa MS AL-Kadmy, Department of Biology, College of Science, AL-Mustansiryiah University, Baghdad-Iraq. Email: [email protected] 1. Definition and Chemical Structure Biopolymer is a term commonly used for polymers which are synthesized by living organisms [1]. Biopolymers originate from natural sources and are biologically renewable, biodegradable and biocompatible. Chitin and chitosan are the biopolymers that have received much research interests due to their numerous potential applications in agriculture, food in- dustry, biomedicine, paper making and textile industry. -

Download (12Mb)
A Thesis Submitted for the Degree of PhD at the University of Warwick Permanent WRAP URL: http://wrap.warwick.ac.uk/110352 Copyright and reuse: This thesis is made available online and is protected by original copyright. Please scroll down to view the document itself. Please refer to the repository record for this item for information to help you to cite it. Our policy information is available from the repository home page. For more information, please contact the WRAP Team at: [email protected] warwick.ac.uk/lib-publications THE BRITISH LIBRARY BRITISH THESIS SERVICE THE DISTRIBUTION OF PHENOTYPIC AND GENOTYPIC CHARACTERS WITHIN STREPTOMYCETES AND THEIR RELATIONSHIP TITLE . TO ANTIBIOTIC PRODUCTION. AUTHOR........ Lesley Phillips, DEGREE.................................................... AWARDING BODY _ TI. .. The University of Warwick, THESIS NUMBER THIS THESIS HAS BEEN MICROFILMED EXACTLY AS RECEIVED The quality of this reproduction is dependent upon the quality of the original thesis submitted for microfilming. Every effort has been made to ensure the highest quality of reproduction. Some pages may have indistinct print, especially if the original papers were poorly produced or if the awarding body sent an inferior copy. If pages are missing, please contact the awarding body which granted the degree. Previously copyrighted materials (journal articles, published texts, etc.) are not filmed. This copy of the thesis has been supplied on condition that anyone who consults it is understood to recognise that Its copyright rests with its author and that no information derived from it may be published without the author's prior written consent. Reproduction of this thesis, other than as permitted under the United Kingdom Copyright Designs and Patents Act 1988, or under specific agreement with the copyright holder, is prohibited. -

Antibiotic Discovery
ANTIBIOTIC DISCOVERY RESISTANCE PROFILING OF MICROBIAL GENOMES TO REVEAL NOVEL ANTIBIOTIC NATURAL PRODUCTS By CHELSEA WALKER, H. BSc. A Thesis Submitted to the School of Graduate Studies in Partial Fulfilment of the Requirements for the Degree Master of Science McMaster University © Copyright by Chelsea Walker, May 2017 McMaster University MASTER OF SCIENCE (2017) Hamilton, Ontario (Biochemistry and Biomedical Sciences) TITLE: Resistance Profiling of Microbial Genomes to Reveal Novel Antibiotic Natural Products. AUTHOR: Chelsea Walker, H. BSc. (McMaster University) SUPERVISOR: Dr. Nathan A. Magarvey. COMMITTEE MEMBERS: Dr. Eric Brown and Dr. Michael G. Surette. NUMBER OF PAGES: xvii, 168 ii Lay Abstract It would be hard to imagine a world where we could no longer use the antibiotics we are routinely being prescribed for common bacterial infections. Currently, we are in an era where this thought could become a reality. Although we have been able to discover antibiotics in the past from soil dwelling microbes, this approach to discovery is being constantly challenged. At the same time, the bacteria are getting smarter in their ways to evade antibiotics, in the form of resistance, or self-protection mechanisms. As such is it essential to devise methods which can predict the potential for resistance to the antibiotics we use early in the discovery and isolation process. By using what we have learned in the past about how bacteria protect themselves for antibiotics, we can to stay one step ahead of them as we continue to search for new sources of antibiotics from bacteria. iii Abstract Microbial natural products have been an invaluable resource for providing clinically relevant therapeutics for almost a century, including most of the commonly used antibiotics that are still in medical use today. -

Study of Actinobacteria and Their Secondary Metabolites from Various Habitats in Indonesia and Deep-Sea of the North Atlantic Ocean
Study of Actinobacteria and their Secondary Metabolites from Various Habitats in Indonesia and Deep-Sea of the North Atlantic Ocean Von der Fakultät für Lebenswissenschaften der Technischen Universität Carolo-Wilhelmina zu Braunschweig zur Erlangung des Grades eines Doktors der Naturwissenschaften (Dr. rer. nat.) genehmigte D i s s e r t a t i o n von Chandra Risdian aus Jakarta / Indonesien 1. Referent: Professor Dr. Michael Steinert 2. Referent: Privatdozent Dr. Joachim M. Wink eingereicht am: 18.12.2019 mündliche Prüfung (Disputation) am: 04.03.2020 Druckjahr 2020 ii Vorveröffentlichungen der Dissertation Teilergebnisse aus dieser Arbeit wurden mit Genehmigung der Fakultät für Lebenswissenschaften, vertreten durch den Mentor der Arbeit, in folgenden Beiträgen vorab veröffentlicht: Publikationen Risdian C, Primahana G, Mozef T, Dewi RT, Ratnakomala S, Lisdiyanti P, and Wink J. Screening of antimicrobial producing Actinobacteria from Enggano Island, Indonesia. AIP Conf Proc 2024(1):020039 (2018). Risdian C, Mozef T, and Wink J. Biosynthesis of polyketides in Streptomyces. Microorganisms 7(5):124 (2019) Posterbeiträge Risdian C, Mozef T, Dewi RT, Primahana G, Lisdiyanti P, Ratnakomala S, Sudarman E, Steinert M, and Wink J. Isolation, characterization, and screening of antibiotic producing Streptomyces spp. collected from soil of Enggano Island, Indonesia. The 7th HIPS Symposium, Saarbrücken, Germany (2017). Risdian C, Ratnakomala S, Lisdiyanti P, Mozef T, and Wink J. Multilocus sequence analysis of Streptomyces sp. SHP 1-2 and related species for phylogenetic and taxonomic studies. The HIPS Symposium, Saarbrücken, Germany (2019). iii Acknowledgements Acknowledgements First and foremost I would like to express my deep gratitude to my mentor PD Dr. -

General Prescription
GENERAL PRESCRIPTION LESSON 1. INTRODUCTION. PRESCRIPTION. SOLID MEDICINAL FORMS Objective: To study the structure of the prescription, learn the rules and get practical skills in writing out solid medicinal forms in prescription. To carry out practical tasks on prescriptions it is recommended to use Appendix 1. Key questions: 1. Pharmacology as a science and the basis of therapy. Main development milestones of modern pharmacology. Sections of Pharmacology. 2. The concept of medicinal substance, medicinal agent (medicinal drug, drug), medicinal form. 3. The concept of the pharmacological action and types of the action of drugs. 4. The sources of obtaining drugs. 5. International and national pharmacopeia, its content and purpose. 6. Pharmacy. Rules of drug storage and dispensing. 7. Prescription and its structure. Prescription forms. General rules for writing out a prescription. State regulation of writing out and dispensing drugs. 8. Name of medicinal products (international non-proprietary name - INN, trade name). 9. Peculiarities of writing out narcotic, poisonous and potent substances in prescription. 10. Drugs under control. Drugs prohibited for prescribing. 11. Solid medicinal forms: tablets, dragee (pills), powders, capsules. Their characteristics, advantages and disadvantages. Rules of prescribing. Write out prescriptions for: 1. 5 powders of Codeine 0.015 g. 1 powder orally twice a day. 2. 10 powders of Didanosine 0.25 g in sachets to prepare solution for internal use. Accept inside twice a day one sachet powder after dissolution in a glass of boiled water. 3. 50 mg of Alteplase powder in the bottle. Dissolve the content of the bottle in 50 ml of saline. First 15 ml introduce intravenously streamly, then intravenous drip. -

Effect of Sulfonylurea Tribenuron Methyl Herbicide on Soil
b r a z i l i a n j o u r n a l o f m i c r o b i o l o g y 4 9 (2 0 1 8) 79–86 ht tp://www.bjmicrobiol.com.br/ Environmental Microbiology Effect of sulfonylurea tribenuron methyl herbicide on soil Actinobacteria growth and characterization of resistant strains a,b,∗ a,c d f e Kounouz Rachedi , Ferial Zermane , Radja Tir , Fatima Ayache , Robert Duran , e e e a,c Béatrice Lauga , Solange Karama , Maryse Simon , Abderrahmane Boulahrouf a Université Frères Mentouri, Faculté des Sciences de la Nature et de la Vie, Laboratoire de Génie Microbiologique et Applications, Constantine, Algeria b Université Frères Mentouri, Institut de la Nutrition, de l’Alimentation et des Technologies Agro-Alimentaires (INATAA), Constantine, Algeria c Université Frères Mentouri, Faculté des Sciences de la Nature et de la Vie, Département de Microbiologie, Constantine, Algeria d Université Frères Mentouri, Faculté des Sciences de la Nature et de la Vie, Laboratoire de Biologie Moléculaire et Cellulaire, Constantine, Algeria e Université de Pau et des Pays de l’Adour, Unité Mixte de Recherche 5254, Equipe Environnement et Microbiologie, Pau, France f Université Frères Mentouri, Constantine 1, Algeria a r t i c l e i n f o a b s t r a c t Article history: Repeated application of pesticides disturbs microbial communities and cause dysfunctions ® Received 28 October 2016 on soil biological processes. Granstar 75 DF is one of the most used sulfonylurea herbi- Accepted 6 May 2017 cides on cereal crops; it contains 75% of tribenuron-methyl.