Genetic Differentiation Between Cave and Surface
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
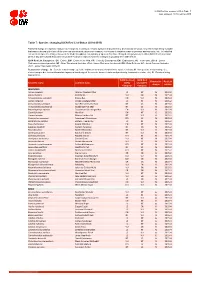
Table 7: Species Changing IUCN Red List Status (2014-2015)
IUCN Red List version 2015.4: Table 7 Last Updated: 19 November 2015 Table 7: Species changing IUCN Red List Status (2014-2015) Published listings of a species' status may change for a variety of reasons (genuine improvement or deterioration in status; new information being available that was not known at the time of the previous assessment; taxonomic changes; corrections to mistakes made in previous assessments, etc. To help Red List users interpret the changes between the Red List updates, a summary of species that have changed category between 2014 (IUCN Red List version 2014.3) and 2015 (IUCN Red List version 2015-4) and the reasons for these changes is provided in the table below. IUCN Red List Categories: EX - Extinct, EW - Extinct in the Wild, CR - Critically Endangered, EN - Endangered, VU - Vulnerable, LR/cd - Lower Risk/conservation dependent, NT - Near Threatened (includes LR/nt - Lower Risk/near threatened), DD - Data Deficient, LC - Least Concern (includes LR/lc - Lower Risk, least concern). Reasons for change: G - Genuine status change (genuine improvement or deterioration in the species' status); N - Non-genuine status change (i.e., status changes due to new information, improved knowledge of the criteria, incorrect data used previously, taxonomic revision, etc.); E - Previous listing was an Error. IUCN Red List IUCN Red Reason for Red List Scientific name Common name (2014) List (2015) change version Category Category MAMMALS Aonyx capensis African Clawless Otter LC NT N 2015-2 Ailurus fulgens Red Panda VU EN N 2015-4 -

§4-71-6.5 LIST of CONDITIONALLY APPROVED ANIMALS November
§4-71-6.5 LIST OF CONDITIONALLY APPROVED ANIMALS November 28, 2006 SCIENTIFIC NAME COMMON NAME INVERTEBRATES PHYLUM Annelida CLASS Oligochaeta ORDER Plesiopora FAMILY Tubificidae Tubifex (all species in genus) worm, tubifex PHYLUM Arthropoda CLASS Crustacea ORDER Anostraca FAMILY Artemiidae Artemia (all species in genus) shrimp, brine ORDER Cladocera FAMILY Daphnidae Daphnia (all species in genus) flea, water ORDER Decapoda FAMILY Atelecyclidae Erimacrus isenbeckii crab, horsehair FAMILY Cancridae Cancer antennarius crab, California rock Cancer anthonyi crab, yellowstone Cancer borealis crab, Jonah Cancer magister crab, dungeness Cancer productus crab, rock (red) FAMILY Geryonidae Geryon affinis crab, golden FAMILY Lithodidae Paralithodes camtschatica crab, Alaskan king FAMILY Majidae Chionocetes bairdi crab, snow Chionocetes opilio crab, snow 1 CONDITIONAL ANIMAL LIST §4-71-6.5 SCIENTIFIC NAME COMMON NAME Chionocetes tanneri crab, snow FAMILY Nephropidae Homarus (all species in genus) lobster, true FAMILY Palaemonidae Macrobrachium lar shrimp, freshwater Macrobrachium rosenbergi prawn, giant long-legged FAMILY Palinuridae Jasus (all species in genus) crayfish, saltwater; lobster Panulirus argus lobster, Atlantic spiny Panulirus longipes femoristriga crayfish, saltwater Panulirus pencillatus lobster, spiny FAMILY Portunidae Callinectes sapidus crab, blue Scylla serrata crab, Samoan; serrate, swimming FAMILY Raninidae Ranina ranina crab, spanner; red frog, Hawaiian CLASS Insecta ORDER Coleoptera FAMILY Tenebrionidae Tenebrio molitor mealworm, -

Labeo Nigripinnis (A Carp, No Common Name) Ecological Risk Screening Summary
Labeo nigripinnis (a carp, no common name) Ecological Risk Screening Summary U.S. Fish and Wildlife Service, May 2012 Revised, May 2018 Web Version, 6/15/2018 1 Native Range and Status in the United States Native Range From Froese and Pauly (2018): “Asia: endemic to Pakistan.” Datta and Majumdar (1970) report L. nigripinnis from the states of Rajasthan and Punjab in northern India, and from western Pakistan. From Sarma et al. (2017): “The present survey work has described 10 species of Labeo sp. [including] Labeo nigripinnis [in Gujarat, India].” From Jayaram and Jeyachandra Dhas (2000): “L. nigripinnis are confined to the Himalayan drainage system and have not spread below the Vindya-Satpura range of mountains [central India].” 1 Status in the United States This species has not been reported as introduced or established in the United States. There is no indication that this species is in trade in the United States. Means of Introductions in the United States This species has not been reported as introduced or established in the United States. Remarks From Jayaram and Jeyachandra Dhas (2000): “L. caeruleus, Labeo diplostomus, L. microphthalmus and L. nigripinnis are all derivatives of L. dero which is found all along the Himalaya including the Sind hills [Pakistan] and Myanmar.” 2 Biology and Ecology Taxonomic Hierarchy and Taxonomic Standing From ITIS (2018): “Kingdom Animalia Subkingdom Bilateria Infrakingdom Deuterostomia Phylum Chordata Subphylum Vertebrata Infraphylum Gnathostomata Superclass Actinopterygii Class Teleostei Superorder -

Monograph of the Cyprinid Fis~Hes of the Genus Garra Hamilton (173)
MONOGRAPH OF THE CYPRINID FIS~HES OF THE GENUS GARRA HAMILTON By A. G. K. MENON, Zoologist, ,Zoological Surt1ey of India, Oalcutta. (With 1 Table, 29 Text-figs. and 6 Plates) CONTENTS Page I-Introduction 175 II-Purpose and general results 176 III-Methods and approaches 176 (a) The definition of Measurements 176 (b) The analysis of Intergradation 178 (c) The recognition of subspecies. 179 (d) Procedures in the paper 180 (e) Evaluation of systematic characters 181 (I) Abbreviations of names of Institutions 181 IV-Historical sketch 182 V-Definition of the genus 187 VI-Systematic section 188 (a) The variabilis group 188 (i) The variabilis Complex 188 1. G. variabilis 188 2. G. rossica 189 (b) The tibanica group 191 (i) The tibanica Complex 191 3. G. tibanica. 191 4. G. quadrimaculata 192 5. G. ignestii 195 6. G. ornata 196 7. G. trewavasi 198 8. G. makiensis 198 9. G. dembeensis 199 10. G. ethelwynnae 202 (ii) The rufa complex 203 11. G. rufa rufa 203 12. G. rufa obtusa 205 13. O. barteimiae 206 (iii) The lamta complex 208 14. G. lamta 208 15. G. mullya 212 16. G. 'ceylonensis ceylonensis 216 17. G. c. phillipsi 216 18. G. annandalei 217 (173) 174 page (iv) The lissorkynckus complex 219 19. G. lissorkynchus 219 20. G. rupecula 220 ~ (v) The taeniata complex 221 21. G. taeniata. 221 22" G. borneensis 224 (vi) The yunnanensis complex 224 23. G. yunnanensis 225 24. G. gracilis 229 25. G. naganensis 226 26. G. kempii 227 27. G. mcOlellandi 228 28. G. -

Safety and Hygiene of Ichthyotherapy with G. Rufa Fish
Arch Phys Glob Res 2019; 23 (2): 37-44 ORIGINAL PAPER DOI 10.15442/apgr.23.2.5 Safety and hygiene of ichthyotherapy with G. rufa fish Małgorzata Gorzel*, Anna Kiełtyka-Dadasiewicz** * Faculty of Health Sciences, Vincent Pol University, ** Research and Science Innovation Center, Lublin Abstract Today, biotherapies (therapies using animals, plants or their secretions) are becoming increasingly popular. In medicine and cosmetology, leeches, fish, insect larvae, mucus fromH. aspersa snail, bee products are used. Among them, ichthyotherapy - therapy with the use of Garra rufa (Heckel 1843), which is gaining popularity not only in the exotic resorts of Turkey, Iran and Jordan, but also in Poland, deserves attention. In view of the growing interest in ichthyotherapy, the question arises: Are cosmetic and therapeutic procedures using this vertebrate completely safe for humans? This question became a contribution to taking up this topic of study. Ichthyotherapy is the use of freshwater, sedentary benthopelagic fish of the cyprinid family, red garra (G. rufa), commercially referred to as “the doctor fish”. This fish has a suction apparatus that allows removal of calloused epidermis in patients undergoing therapy. This treatment has been used mainly in cosmetics as so-called fish pedicure, but also in medicine. Scientific reports indicate that this fish may be helpful in treating some skin diseases, i.e. in alleviating the symptoms of psoriasis or atopic dermatitis. Treatments using the red garra have to be performed in full compliance with hygiene rules. In 2011, the British Health Protect Agency (BHPA) published the guidelines for carrying out treatments using these fish. It specified the indications and contraindications for performing the procedure, the way it should be performed, as well as the threats that could result from possible non-compliance with the principles of occupational health and safety during the procedures. -

A Review of Freshwater Fish Fauna in Jordan
Libyan Agriculture Research Center Journal International 1 (5): 320-324, 2010 ISSN 2219-4304 © IDOSI Publications, 2010 A Review of Freshwater Fish Fauna in Jordan Saleh Ahmad Al-Quran Department of Biology, Faculty of Science, Mutah University, Karak, Jordan Abstract: A total of 27 fish species belonging to ten families were recorded. The fish fauna distribution in two rivers (Jordan and Yarmouk) and some dams and tributaries in Jordan was examined. The rivers surveyed were generally shallow, fast flowing with clear water and rocky and sandy substrate. At the time of survey, the rivers gave excellent water quality data. Cyprinidae has the highest diversity 0.296% with 8 species; while the lowest one are Anguillidae, Blennidae, Clariidae, Mugilidae and Pseudocrenilabrinae; each represented only by one species (0.037%). Cichlidae represented by 6 species (0.222%), Balitoridae and Cyprinodontidae are represented by three species (0.111%) and finally Acanthuridae is represented by two species (0.074%). Key words: Review % Freshwater % Fish % Fauna % Jordan INTRODUCTION drains into the Dead Sea which, at 407 meters below sea level, is the lowest point on earth. The river is between Location, Topography and Climate: Jordan is a relatively 20 and 30 meters wide near its endpoint. Its flow has been small country situated at the junction of the Levantine much reduced and its salinity increased because and Arabian areas of the Middle East. The country is significant amounts have been diverted for irrigational bordered on the north by Syria, to the east by Iraq and by uses. Several degrees warmer than the rest of the country, Saudi Arabia on the east and south. -

Genetic Diversity and Population Structure of the Critically Endangered Freshwater Fish Species, the Clanwilliam Sandfish (Labeo Seeberi)
Genetic Diversity and Population Structure of the critically endangered freshwater fish species, the Clanwilliam sandfish (Labeo seeberi) By Shaun Francois Lesch Thesis presented in partial fulfilment of the requirements for the degree of Master of Science in the Faculty of Natural Science at Stellenbosch University Supervisor: Dr C. Rhode Co-supervisor: Dr R. Slabbert Department of Genetics December 2020 Stellenbosch University https://scholar.sun.ac.za Declaration: By submitting this thesis electronically, I declare that the entirety of the work contained therein is my own, original work, that I am the sole author thereof (save to the extent explicitly otherwise stated), that reproduction and publication thereof by Stellenbosch University will not infringe any third party rights and that I have not previously in its entirety or in part submitted it for obtaining any qualification. Date: December 2020 Copyright © 2020 Stellenbosch University All Rights Reserved i Stellenbosch University https://scholar.sun.ac.za Abstract: Labeo spp. are large freshwater fish found throughout southern Asia, the Middle East and Africa. The genus is characterised by specialised structures around the mouth and lips making it adapted to herbivorous feeding (algae and detritus). Clanwilliam sandfish (Labeo seeberi) was once widespread throughout its natural habitat (Olifants-Doring River system), but significant decreases in population size have seen them become absent in the Olifants River and retreat to the headwaters in the tributaries of the Doring River. Currently sandfish are confined to three populations namely the Oorlogskloof Nature Reserve (OKNR), Rietkuil (Riet) and Bos, with OKNR being the largest of the three and deemed the species sanctuary. -

Morphological and Molecular Studies on Garra Imberba and Its Related Species in China
Zoological Research 35 (1): 20−32 DOI:10.11813/j.issn.0254-5853.2014.1.020 Morphological and molecular studies on Garra imberba and its related species in China Wei-Ying WANG 1,2, Wei ZHOU3, Jun-Xing YANG1, Xiao-Yong CHEN1,* 1. State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China 2. University of Chinese Academy of Sciences, Beijing 100049, China 3. Southwest Forestry University, Kunming, Yunnan 650224, China Abstract: Garra imberba is widely distributed in China. At the moment, both Garra yiliangensis and G. hainanensis are treated as valid species, but they were initially named as a subspecies of G. pingi, a junior synonym of G. imberba. Garra alticorpora and G. nujiangensis also have similar morphological characters to G. imberba, but the taxonomic statuses and phylogenetic relationships of these species with G. imberba remains uncertain. In this study, 128 samples from the Jinshajiang, Red, Nanpanjiang, Lancangjiang, Nujiang Rivers as well as Hainan Island were measured while 1 mitochondrial gene and 1 nuclear intron of 24 samples were sequenced to explore the phylogenetic relationship of these five species. The results showed that G. hainanensis, G. yiliangensis, G. alticorpora and G. imberba are the same species with G. imberba being the valid species name, while G. nujiangensis is a valid species in and of itself. Keywords: Garra imberba; Taxonomy; Morphology; Molecular phylogeny With 105 valid species Garra is one of the most from Jinshajiang River as G. pingi, but Kottelat (1998) diverse genera of the Labeoninae, and has a widespread treated G. -

Review of Freshwater Fish
CMS Distribution: General CONVENTION ON MIGRATORY UNEP/CMS/Inf.10.33 1 November 2011 SPECIES Original: English TENTH MEETING OF THE CONFERENCE OF THE PARTIES Bergen, 20-25 November 2011 Agenda Item 19 REVIEW OF FRESHWATER FISH (Prepared by Dr. Zeb Hogan, COP Appointed Councillor for Fish) Pursuant to the Strategic Plan 2006-2011 mandating a review of the conservation status for Appendix I and II species at regular intervals, the 15 th Meeting of the Scientific Council (Rome, 2008) tasked the COP Appointed Councillor for Fish, Mr. Zeb Hogan, with preparing a report on the conservation status of CMS-listed freshwater fish. The report, which reviews available population assessments and provides guidance for including further freshwater fish on the CMS Appendices, is presented in this Information Document in the original form in which it was delivered to the Secretariat. Preliminary results were discussed at the 16 th Meeting of the Scientific Council (Bonn, 2010). An executive summary is provided as document UNEP/CMS/Conf.10.31 and a Resolution as document UNEP/CMS/Resolution 10.12. For reasons of economy, documents are printed in a limited number, and will not be distributed at the meeting. Delegates are kindly requested to bring their copy to the meeting and not to request additional copies. Review of Migratory Freshwater Fish Prepared by Dr. Zeb Hogan, CMS Scientific Councilor for Fish on behalf of the CMS Secretariat 1 Table of Contents Acknowledgements .................................................................................................................................3 -

Adding Nuclear Rhodopsin Data Where Mitochondrial COI Indicates Discrepancies – Can This Marker Help to Explain Conflicts in Cyprinids?
DNA Barcodes 2015; 3: 187–199 Research Article Open Access S. Behrens-Chapuis*, F. Herder, H. R. Esmaeili, J. Freyhof, N. A. Hamidan, M. Özuluğ, R. Šanda, M. F. Geiger Adding nuclear rhodopsin data where mitochondrial COI indicates discrepancies – can this marker help to explain conflicts in cyprinids? DOI 10.1515/dna-2015-0020 or haplotype sharing (12 species pairs) with presumed Received February 27, 2015; accepted October 1, 2015 introgression based on mtCOI data. We aimed to test Abstract: DNA barcoding is a fast and reliable tool for the utility of the nuclear rhodopsin marker to uncover species identification, and has been successfully applied reasons for the high similarity and haplotype sharing in to a wide range of freshwater fishes. The limitations these different groups. The included labeonine species reported were mainly attributed to effects of geographic belonging to Crossocheilus, Hemigrammocapoeta, scale, taxon-sampling, incomplete lineage sorting, or Tylognathus and Typhlogarra were found to be nested mitochondrial introgression. However, the metrics for within the genus Garra based on mtCOI. This specific the success of assigning unknown samples to species or taxonomic uncertainty was also addressed by the use genera also depend on a suited taxonomic framework. of the additional nuclear marker. As a measure of the A simultaneous use of the mitochondrial COI and the delineation success we computed barcode gaps, which nuclear RHO gene turned out to be advantageous for were present in 75% of the species based on mtCOI, but the barcode efficiency in a few previous studies. Here, in only 39% based on nuclear rhodopsin sequences. -

Fish-Released Kairomones Affect Culiseta Longiareolata Oviposition and Larval Life History
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.22.423951; this version posted December 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Fish-released kairomones affect Culiseta longiareolata oviposition and larval life history Alon Silberbush Department of Biology and the Environment, Faculty of Natural Sciences, University of Haifa, Israel Abstract: Several species of mosquitoes respond to the presence of kairomones released by larval predators during oviposition habitat selection and larval development. These responses may differ among mosquito species and do not always correlate with larval survival. This study examined the responses of the mosquito Culiseta longiareolata Macquart to kairomones released by three species of fish during oviposition, Gambusia affinis Baird and Girard, Aphanius mento Heckel and Garra rufa Heckel. In addition, the study examined the effects of kairomones released by G. affinis on larval development. Results show that ovipositing female avoided cues from larvivorous, but not algivorous fish. In addition, developing larvae metamorphosed slower and showed increased mortality when exposed to fish-released kairomones. Results suggest that the responses of this mosquito species to fish-released kairomones may be explained by its competitive ability. Keywords: Predator-released kairomones, life history, oviposition habitat selection. Introduction: Semiochemicals indicating the presence of predators are often an important part of predator detection mechanisms in addition to visual, audio and tactical cues (Kats and Dill 1998). Aquatic organisms often rely specifically on kairomones or other predator associated semiochemicals, bioRxiv preprint doi: https://doi.org/10.1101/2020.12.22.423951; this version posted December 22, 2020. -

Training Manual Series No.15/2018
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by CMFRI Digital Repository DBTR-H D Indian Council of Agricultural Research Ministry of Science and Technology Central Marine Fisheries Research Institute Department of Biotechnology CMFRI Training Manual Series No.15/2018 Training Manual In the frame work of the project: DBT sponsored Three Months National Training in Molecular Biology and Biotechnology for Fisheries Professionals 2015-18 Training Manual In the frame work of the project: DBT sponsored Three Months National Training in Molecular Biology and Biotechnology for Fisheries Professionals 2015-18 Training Manual This is a limited edition of the CMFRI Training Manual provided to participants of the “DBT sponsored Three Months National Training in Molecular Biology and Biotechnology for Fisheries Professionals” organized by the Marine Biotechnology Division of Central Marine Fisheries Research Institute (CMFRI), from 2nd February 2015 - 31st March 2018. Principal Investigator Dr. P. Vijayagopal Compiled & Edited by Dr. P. Vijayagopal Dr. Reynold Peter Assisted by Aditya Prabhakar Swetha Dhamodharan P V ISBN 978-93-82263-24-1 CMFRI Training Manual Series No.15/2018 Published by Dr A Gopalakrishnan Director, Central Marine Fisheries Research Institute (ICAR-CMFRI) Central Marine Fisheries Research Institute PB.No:1603, Ernakulam North P.O, Kochi-682018, India. 2 Foreword Central Marine Fisheries Research Institute (CMFRI), Kochi along with CIFE, Mumbai and CIFA, Bhubaneswar within the Indian Council of Agricultural Research (ICAR) and Department of Biotechnology of Government of India organized a series of training programs entitled “DBT sponsored Three Months National Training in Molecular Biology and Biotechnology for Fisheries Professionals”.