Adding Nuclear Rhodopsin Data Where Mitochondrial COI Indicates Discrepancies – Can This Marker Help to Explain Conflicts in Cyprinids?
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
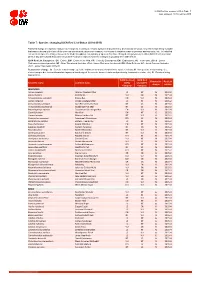
Table 7: Species Changing IUCN Red List Status (2014-2015)
IUCN Red List version 2015.4: Table 7 Last Updated: 19 November 2015 Table 7: Species changing IUCN Red List Status (2014-2015) Published listings of a species' status may change for a variety of reasons (genuine improvement or deterioration in status; new information being available that was not known at the time of the previous assessment; taxonomic changes; corrections to mistakes made in previous assessments, etc. To help Red List users interpret the changes between the Red List updates, a summary of species that have changed category between 2014 (IUCN Red List version 2014.3) and 2015 (IUCN Red List version 2015-4) and the reasons for these changes is provided in the table below. IUCN Red List Categories: EX - Extinct, EW - Extinct in the Wild, CR - Critically Endangered, EN - Endangered, VU - Vulnerable, LR/cd - Lower Risk/conservation dependent, NT - Near Threatened (includes LR/nt - Lower Risk/near threatened), DD - Data Deficient, LC - Least Concern (includes LR/lc - Lower Risk, least concern). Reasons for change: G - Genuine status change (genuine improvement or deterioration in the species' status); N - Non-genuine status change (i.e., status changes due to new information, improved knowledge of the criteria, incorrect data used previously, taxonomic revision, etc.); E - Previous listing was an Error. IUCN Red List IUCN Red Reason for Red List Scientific name Common name (2014) List (2015) change version Category Category MAMMALS Aonyx capensis African Clawless Otter LC NT N 2015-2 Ailurus fulgens Red Panda VU EN N 2015-4 -

Review and Updated Checklist of Freshwater Fishes of Iran: Taxonomy, Distribution and Conservation Status
Iran. J. Ichthyol. (March 2017), 4(Suppl. 1): 1–114 Received: October 18, 2016 © 2017 Iranian Society of Ichthyology Accepted: February 30, 2017 P-ISSN: 2383-1561; E-ISSN: 2383-0964 doi: 10.7508/iji.2017 http://www.ijichthyol.org Review and updated checklist of freshwater fishes of Iran: Taxonomy, distribution and conservation status Hamid Reza ESMAEILI1*, Hamidreza MEHRABAN1, Keivan ABBASI2, Yazdan KEIVANY3, Brian W. COAD4 1Ichthyology and Molecular Systematics Research Laboratory, Zoology Section, Department of Biology, College of Sciences, Shiraz University, Shiraz, Iran 2Inland Waters Aquaculture Research Center. Iranian Fisheries Sciences Research Institute. Agricultural Research, Education and Extension Organization, Bandar Anzali, Iran 3Department of Natural Resources (Fisheries Division), Isfahan University of Technology, Isfahan 84156-83111, Iran 4Canadian Museum of Nature, Ottawa, Ontario, K1P 6P4 Canada *Email: [email protected] Abstract: This checklist aims to reviews and summarize the results of the systematic and zoogeographical research on the Iranian inland ichthyofauna that has been carried out for more than 200 years. Since the work of J.J. Heckel (1846-1849), the number of valid species has increased significantly and the systematic status of many of the species has changed, and reorganization and updating of the published information has become essential. Here we take the opportunity to provide a new and updated checklist of freshwater fishes of Iran based on literature and taxon occurrence data obtained from natural history and new fish collections. This article lists 288 species in 107 genera, 28 families, 22 orders and 3 classes reported from different Iranian basins. However, presence of 23 reported species in Iranian waters needs confirmation by specimens. -

Scale Deformities in Three Species of the Genus Garra (Actinopterygii: Cyprinidae)
Scale deformities in three species of the genus Garra (Actinopterygii: Cyprinidae) Halimeh Zareian1,2, Hamid Reza Esmaeili1, Ali Gholamhosseini1* 1. Developmental Biosystematics Research Laboratory, Zoology Section, Department of Biology, School of Science, Shiraz University, Shiraz, Iran 2. Zand Institute of Higher Education, Shiraz, Iran *Corresponding author’s E-mail: [email protected] ABSTRACT Different types of scale deformities have been reported from fishes worldwide, however there is no available study on the abnormal scales in the genus Garra except for G. variabilis. In the present study, scale deformities of three species of Garra including G. rufa, G. persica and Garra sp. from 6 sites of the Iranian drainages were examined and described. Different deformations were observed in focus, anterior, posterior and lateral sides of scales in the studied species, showing both slight and severe abnormalities. The occurrence of twin scales was one of the most interesting cases among various types of scale deformities observed on G. persica and Garra sp. Genetic disorders, diseases (including infection and lesions), developmental anomalies, incomplete regeneration after wounding, physical, and chemical environmental variables including pollutions might be considered as potential factors for scale abnormalities remained to be investigated. Keywords: Garra, Scale morphology, Taxonomy, Abnormal scale, Iranian drainage basins. INTRODUCTION Among the morphological abnormalities reported in fishes (e.g. Poppe et al. 1997; Corrales -

Chondrostoma Nasus) Ecological Risk Screening Summary
Common Nase (Chondrostoma nasus) Ecological Risk Screening Summary U.S. Fish & Wildlife Service, April 2020 Revised, April 2020 Web Version, 2/8/2021 Organism Type: Fish Overall Risk Assessment Category: High Photo: André Karwath. Licensed under CC BY-SA 2.5. Available: https://commons.wikimedia.org/wiki/File:Chondrostoma_nasus_(aka).jpg#file. (April 2020). 1 Native Range and Status in the United States Native Range From Froese and Pauly (2021): “Europe: Basins of Black (Danube, Dniestr, South Bug and Dniepr drainages), southern Baltic (Nieman, Odra, Vistula) and southern North Seas (westward to Meuse). […] Asia: Turkey.” Status in the United States No information on occurrence, status, sale or trade in the United States was found. Chondrostoma nasus falls within Group I of New Mexico’s Department of Game and Fish Director’s Species Importation List (New Mexico Department of Game and Fish 2010). Group I species “are designated semi-domesticated animals and do not require an importation permit.” With the added restriction of “Not to be used as bait fish.” 1 Means of Introductions in the United States No introductions have been reported in the United States. Remarks Although the accepted and most used common name for Chondrostoma nasus is “Common Nase”, it appears that the simple name “Nase” is sometimes used to refer to C. nasus (Zbinden and Maier 1996; Jirsa et al. 2010). The name “Sneep” also occasionally appears in the literature (Irz et al. 2006). 2 Biology and Ecology Taxonomic Hierarchy and Taxonomic Standing From Fricke et al. (2020): -

Monograph of the Cyprinid Fis~Hes of the Genus Garra Hamilton (173)
MONOGRAPH OF THE CYPRINID FIS~HES OF THE GENUS GARRA HAMILTON By A. G. K. MENON, Zoologist, ,Zoological Surt1ey of India, Oalcutta. (With 1 Table, 29 Text-figs. and 6 Plates) CONTENTS Page I-Introduction 175 II-Purpose and general results 176 III-Methods and approaches 176 (a) The definition of Measurements 176 (b) The analysis of Intergradation 178 (c) The recognition of subspecies. 179 (d) Procedures in the paper 180 (e) Evaluation of systematic characters 181 (I) Abbreviations of names of Institutions 181 IV-Historical sketch 182 V-Definition of the genus 187 VI-Systematic section 188 (a) The variabilis group 188 (i) The variabilis Complex 188 1. G. variabilis 188 2. G. rossica 189 (b) The tibanica group 191 (i) The tibanica Complex 191 3. G. tibanica. 191 4. G. quadrimaculata 192 5. G. ignestii 195 6. G. ornata 196 7. G. trewavasi 198 8. G. makiensis 198 9. G. dembeensis 199 10. G. ethelwynnae 202 (ii) The rufa complex 203 11. G. rufa rufa 203 12. G. rufa obtusa 205 13. O. barteimiae 206 (iii) The lamta complex 208 14. G. lamta 208 15. G. mullya 212 16. G. 'ceylonensis ceylonensis 216 17. G. c. phillipsi 216 18. G. annandalei 217 (173) 174 page (iv) The lissorkynckus complex 219 19. G. lissorkynchus 219 20. G. rupecula 220 ~ (v) The taeniata complex 221 21. G. taeniata. 221 22" G. borneensis 224 (vi) The yunnanensis complex 224 23. G. yunnanensis 225 24. G. gracilis 229 25. G. naganensis 226 26. G. kempii 227 27. G. mcOlellandi 228 28. G. -

Chondrostoma Soetta Region: 1 Taxonomic Authority: Bonaparte, 1840 Synonyms: Common Names: Savetta Italian Order: Cypriniformes Family: Cyprinidae Notes on Taxonomy
Chondrostoma soetta Region: 1 Taxonomic Authority: Bonaparte, 1840 Synonyms: Common Names: Savetta Italian Order: Cypriniformes Family: Cyprinidae Notes on taxonomy: General Information Biome Terrestrial Freshwater Marine Geographic Range of species: Habitat and Ecology Information: Restricted to northern Italy, the southern part of Switzerland. It has A deepwater lacustrine species that also inhabits large rivers. It been introduced in some Italian lakes. It is locally extinct in Slovenia migrates from the lake to its tributaries for spawning in spring. and the Isonzo river basin in Italy due to the introduction of Chondrostoma nasus, a practice still implemented. Introductions in rivers of central Italy was often a misidentification for C. genei. Several present records for C. soetta are probably a misidentification for C. nasus, due to similarity between these two species. As an example of how the alien species are spread now in Italy, in rivers from the Rovigo Province in eastern Italy, were C. soetta is still reported, the biomass of all native species was found to be about 22% of whole ichthyofauna. Conservation Measures: Threats: Listed in Annex II of the Habitats Directive of EU and in the Appendix III Dams, water pollution and extraction, and introduction of alien species of the Bern Convention. as Rutilus rutilus, Silurus glanis and Chondrostoma nasus. The reduction in suitable spawning places due to pollution (agriculture) and to water extraction is of major concern. Other threats to the species are predation by cormorants, where in several places of Italy have become a serious pest and destroyed a large amount of fishes, especially in torrents or small river were the fishes migrate to for reproduction. -

Safety and Hygiene of Ichthyotherapy with G. Rufa Fish
Arch Phys Glob Res 2019; 23 (2): 37-44 ORIGINAL PAPER DOI 10.15442/apgr.23.2.5 Safety and hygiene of ichthyotherapy with G. rufa fish Małgorzata Gorzel*, Anna Kiełtyka-Dadasiewicz** * Faculty of Health Sciences, Vincent Pol University, ** Research and Science Innovation Center, Lublin Abstract Today, biotherapies (therapies using animals, plants or their secretions) are becoming increasingly popular. In medicine and cosmetology, leeches, fish, insect larvae, mucus fromH. aspersa snail, bee products are used. Among them, ichthyotherapy - therapy with the use of Garra rufa (Heckel 1843), which is gaining popularity not only in the exotic resorts of Turkey, Iran and Jordan, but also in Poland, deserves attention. In view of the growing interest in ichthyotherapy, the question arises: Are cosmetic and therapeutic procedures using this vertebrate completely safe for humans? This question became a contribution to taking up this topic of study. Ichthyotherapy is the use of freshwater, sedentary benthopelagic fish of the cyprinid family, red garra (G. rufa), commercially referred to as “the doctor fish”. This fish has a suction apparatus that allows removal of calloused epidermis in patients undergoing therapy. This treatment has been used mainly in cosmetics as so-called fish pedicure, but also in medicine. Scientific reports indicate that this fish may be helpful in treating some skin diseases, i.e. in alleviating the symptoms of psoriasis or atopic dermatitis. Treatments using the red garra have to be performed in full compliance with hygiene rules. In 2011, the British Health Protect Agency (BHPA) published the guidelines for carrying out treatments using these fish. It specified the indications and contraindications for performing the procedure, the way it should be performed, as well as the threats that could result from possible non-compliance with the principles of occupational health and safety during the procedures. -

Reassessment of the Taxonomic Position of Iranocypris Typhlops Bruun & Kaiser, 1944 (Actinopterygii, Cyprinidae)
A peer-reviewed open-access journal ZooKeysReassessment 374: 69–77 (2014) of the taxonomic position of Iranocypris typhlops Bruun and Kaiser, 1944... 69 doi: 10.3897/zookeys.374.6617 RESEARCH ARTICLE www.zookeys.org Launched to accelerate biodiversity research Reassessment of the taxonomic position of Iranocypris typhlops Bruun & Kaiser, 1944 (Actinopterygii, Cyprinidae) Azita Farashi1, Mohammad Kaboli1, Hamid Reza Rezaei2, Mohammad Reza Naghavi3, Hassan Rahimian4, Brian W. Coad5 1 Department of Environmental Sciences, Faculty of Natural Resources, University of Tehran, Iran 2 Department of Environmental Sciences, Faculty of Natural Resources, Gorgan University, Iran 3 Department of Agronomy and Plant Breeding, Faculty of Agricultural Sciences, University of Tehran, Iran 4 Department of Animal Bio- logy, Faculty of Biology, University of Tehran, Iran 5 Canadian Museum of Nature, Ottawa, Ontario, Canada Corresponding author: Mohammad Kaboli ([email protected]) Academic editor: K. Jordaens | Received 15 November 2013 | Accepted 9 January 2014 | Published 29 January 2014 Citation: Farashi A, Kaboli M, Rezaei HR, Naghavi MR, Rahimian H, Coad BW (2014) Reassessment of the taxonomic position of Iranocypris typhlops Bruun & Kaiser, 1944 (Actinopterygii, Cyprinidae). ZooKeys 374: 69–77. doi: 10.3897/ zookeys.374.6617 Abstract The Iranian cave barb (Iranocypris typhlops Bruun & Kaiser, 1944) is a rare and endemic species of the family Cyprinidae known from a single locality in the Zagros Mountains, western Iran. This species is “Vulnerable” according to the IUCN Red List and is one of the top four threatened freshwater fish species in Iran. Yet, the taxonomic position of I. typhlops is uncertain. We examined phylogenetic relationships of this species with other species of the family Cyprinidae based on the mitochondrial cytochrome b gene. -

Eurasian Minnow (Phoxinus Phoxinus) ERSS
Eurasian Minnow (Phoxinus phoxinus) Ecological Risk Screening Summary U.S. Fish & Wildlife Service, August 2012 Revised, February 2019 Web Version, 8/28/2019 Photo: J. Renoult. Licensed under CC BY-SA 4.0 International. Available: https://api.gbif.org/v1/image/unsafe/https%3A%2F%2Fstatic.inaturalist.org%2Fphotos%2F1624 9424%2Foriginal.jpeg%3F1524597277. (March 2019) 1 Native Range and Status in the United States Native Range From Froese and Pauly (2019): “Eurasia: basins of Atlantic, North and Baltic Seas, Arctic and northern Pacific Ocean, from Garonne (France) eastward to Anadyr and Amur drainages and Korea; Ireland (possibly introduced), Great Britain northward to 58°N. Scandinavia and Russia northernmost extremity, Rhône drainage. Recorded from upper and middle Volga and Ural drainages, Lake Balkhash 1 (Kazakhstan) and upper Syr-Darya drainage (Aral basin), but else identifications need verification.” From GISD (2019): “Albania, Armenia, Azerbaijan, Belarus, Belgium, Bosnia And Herzegovina, Bulgaria, China, Croatia, Czech Republic, Denmark, Estonia, Finland, France, Georgia, Germany, Greece, Hungary, Italy, Kazakhstan, Korea, Democratic People's Republic Of Korea, Republic Of Latvia, Liechtenstein, Lithuania, Luxembourg, Macedonia, The Former Yugoslav Republic Of Moldova, Republic Of Mongolia, Montenegro, Netherlands, Norway, Poland, Romania, Russian Federation, Serbia, Slovakia, Slovenia, Spain, Sweden, Switzerland, Turkey, Ukraine, and the United Kingdom.” In addition to the above locations, Freyhof and Kottelat (2008) lists Andorra and Austria as locations where Phoxinus phoxinus is native. CABI (2019) lists Phoxinus phoxinus as localized in Egypt. Status in the United States No records of introductions of Phoxinus phoxinus in the United States were found. Phoxinus phoxinus was officially listed as an injurious wildlife species in 2016 under the Lacey Act (18.U.S.C.42(a)(1)) by the U.S. -

English Enhancing Biodiversity by Restoring Source Areas for Priority and Other Species of Community Interest in Ticino Park
LIFE15 NAT/IT/000989 N. 2 JUNE2020 News letterEnglish Enhancing Biodiversity by Restoring Source Areas for Priority and Other Species of Community Interest in Ticino Park LIFE Ticino Biosource is a 4 year long project (total budget: 3.877.000 euros) for the conservation of biodiversity in Ticino Park, co- a first assessment three funded mainly by the European Commission, with the contribution of Ticino Park, Fondazione years after the beginning Cariplo, GRAIA and Fondazione Lombardia per l’Ambiente, the last two being as well partners of of the project the project. The project started in October 2016 and will finish in September 2020. On October 4, 2019, a workshop was held at the Park Centre “La Fagiana”, organized by the Ticino Park as part of the E2 Action, entitled “Life Projects to Protect Fish Fauna”. The aim was to create an opportunity to meet, knowledge, information and exchange of good practices and expertise among technicians, researchers and administrators engaged in conservation actions that are similar to those undertaken within the BIOSOURCE project or engaged in LIFE projects that share an interest in the same target species or habitats. The workshop was attended as speakers by 9 representatives of European projects (LIFE or INTERREG projects) which were being implemented, recently completed or even at the proposal stage. On this occasion, in addition to the Life Ticino BIOSOURCE, the following projects were illustrated: - LIFE-Nature project “IdroLIFE”, presented by the project manager Pietro Volta, researcher at the CNR-IRSA; - LIFE-Nature project “LIFE for LASCA”, illustrated by the project manager Kaja Pliberek, from the Fisheries Research Institute of Slovenia - INTERREG project “SHARESALMO”, presented by 1 That’s why, IdroLIFE aims to improve the LIFE15 NAT/IT/000823 conservation status of the local populations of these species in Natura 2000 sites of Verbano Cusio IdroLIFE Ossola Province, contributing to halt the loss of Conservation and management of freshwater fauna aquatic biodiversity. -

A New Record of Iranian Subterranean Fishes Reveals the Potential Presence of a Large Freshwater Aquifer in the Zagros Mountains
Received: 13 April 2019 | Revised: 15 July 2019 | Accepted: 31 July 2019 DOI: 10.1111/jai.13964 ORIGINAL ARTICLE A new record of Iranian subterranean fishes reveals the potential presence of a large freshwater aquifer in the Zagros Mountains Saber Vatandoust1 | Hamed Mousavi‐Sabet2,3 | Matthias F. Geiger4 | Jörg Freyhof5 1Department of Fisheries, Babol Branch, Islamic Azad University, Babol, Iran Abstract 2Department of Fisheries, Faculty of Natural A new locality is reported for the Iranian subterranean fishes Garra typhlops and Resources, University of Guilan, Sowmeh Garra lorestanensis (and probably Eidinemacheilus smithi), near the village Tuveh in the Sara, Iran 3The Caspian Sea Basin Research Dez River drainage. The site is 31 km straight‐line distance away from the only other Center, University of Guilan, Rasht, Iran known locality where these species have been observed previously. The finding sug‐ 4 Zoological Research Museum Alexander gests the presence of a sizeable subterranean aquifer system in the Tigris drainage Koenig, Leibniz Institute for Animal Biodiversity, Bonn, Germany extending for between 31 and 162 km. 5Museum für Naturkunde, Leibniz Institute for Evolution and Biodiversity Science, KEYWORDS Berlin, Germany cyprinidae, cytochrome oxidase i, distribution, freshwater fish Correspondence Hamed Mousavi‐Sabet, Department of Fisheries, Faculty of Natural Resources, University of Guilan, Sowmeh Sara, P.O. Box: 1144, Guilan, Iran. Email: [email protected] 1 | INTRODUCTION Loven (Figure 2) and it is the aim of this study -

Adding Nuclear Rhodopsin Data Where Mitochondrial COI Indicates Discrepancies – Can This Marker Help to Explain Conflicts in Cyprinids?
DNA Barcodes 2015; 3: 187–199 Research Article Open Access S. Behrens-Chapuis*, F. Herder, H. R. Esmaeili, J. Freyhof, N. A. Hamidan, M. Özuluğ, R. Šanda, M. F. Geiger Adding nuclear rhodopsin data where mitochondrial COI indicates discrepancies – can this marker help to explain conflicts in cyprinids? DOI 10.1515/dna-2015-0020 or haplotype sharing (12 species pairs) with presumed Received February 27, 2015; accepted October 1, 2015 introgression based on mtCOI data. We aimed to test Abstract: DNA barcoding is a fast and reliable tool for the utility of the nuclear rhodopsin marker to uncover species identification, and has been successfully applied reasons for the high similarity and haplotype sharing in to a wide range of freshwater fishes. The limitations these different groups. The included labeonine species reported were mainly attributed to effects of geographic belonging to Crossocheilus, Hemigrammocapoeta, scale, taxon-sampling, incomplete lineage sorting, or Tylognathus and Typhlogarra were found to be nested mitochondrial introgression. However, the metrics for within the genus Garra based on mtCOI. This specific the success of assigning unknown samples to species or taxonomic uncertainty was also addressed by the use genera also depend on a suited taxonomic framework. of the additional nuclear marker. As a measure of the A simultaneous use of the mitochondrial COI and the delineation success we computed barcode gaps, which nuclear RHO gene turned out to be advantageous for were present in 75% of the species based on mtCOI, but the barcode efficiency in a few previous studies. Here, in only 39% based on nuclear rhodopsin sequences.