Histone Chaperone CHAF1A Inhibits Differentiation and Promotes Aggressive Neuroblastoma
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Regulation of Oxidized Base Damage Repair by Chromatin Assembly Factor 1 Subunit a Chunying Yang1,*,†, Shiladitya Sengupta1,2,*,†, Pavana M
Published online 27 October 2016 Nucleic Acids Research, 2017, Vol. 45, No. 2 739–748 doi: 10.1093/nar/gkw1024 Regulation of oxidized base damage repair by chromatin assembly factor 1 subunit A Chunying Yang1,*,†, Shiladitya Sengupta1,2,*,†, Pavana M. Hegde1,JoyMitra1, Shuai Jiang3, Brooke Holey3, Altaf H. Sarker3, Miaw-Sheue Tsai3, Muralidhar L. Hegde1,2,4 and Sankar Mitra1,2,* 1Department of Radiation Oncology, Houston Methodist Research Institute, Houston, TX 77030, USA, 2Weill Cornell Medical College, Cornell University, New York, NY 10065, USA, 3Biological Systems and Engineering Division, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA and 4Houston Methodist Neurological Institute, Houston, TX 77030, USA Received March 23, 2016; Revised October 13, 2016; Editorial Decision October 17, 2016; Accepted October 19, 2016 ABSTRACT INTRODUCTION Reactive oxygen species (ROS), generated both en- ROS, continuously generated in mammalian cells both en- dogenously and in response to exogenous stress, in- dogenously and by environmental genotoxicants, induce duce point mutations by mis-replication of oxidized various genomic lesions, including oxidized bases, abasic bases and other lesions in the genome. Repair of (AP) sites and single-strand breaks (SSBs). If unrepaired or these lesions via base excision repair (BER) pathway mis-repaired, DNA lesions would cause mutations which may lead to cytotoxicity and cell death and also carcino- maintains genomic fidelity. Regulation of the BER genic transformation (1). The base excision repair (BER) pathway for mutagenic oxidized bases, initiated by pathway, responsible for repair of oxidized base lesions NEIL1 and other DNA glycosylases at the chromatin which contribute to drug/radiation sensitivity is highly con- level remains unexplored. -

Supplementary Data
SUPPLEMENTARY DATA A cyclin D1-dependent transcriptional program predicts clinical outcome in mantle cell lymphoma Santiago Demajo et al. 1 SUPPLEMENTARY DATA INDEX Supplementary Methods p. 3 Supplementary References p. 8 Supplementary Tables (S1 to S5) p. 9 Supplementary Figures (S1 to S15) p. 17 2 SUPPLEMENTARY METHODS Western blot, immunoprecipitation, and qRT-PCR Western blot (WB) analysis was performed as previously described (1), using cyclin D1 (Santa Cruz Biotechnology, sc-753, RRID:AB_2070433) and tubulin (Sigma-Aldrich, T5168, RRID:AB_477579) antibodies. Co-immunoprecipitation assays were performed as described before (2), using cyclin D1 antibody (Santa Cruz Biotechnology, sc-8396, RRID:AB_627344) or control IgG (Santa Cruz Biotechnology, sc-2025, RRID:AB_737182) followed by protein G- magnetic beads (Invitrogen) incubation and elution with Glycine 100mM pH=2.5. Co-IP experiments were performed within five weeks after cell thawing. Cyclin D1 (Santa Cruz Biotechnology, sc-753), E2F4 (Bethyl, A302-134A, RRID:AB_1720353), FOXM1 (Santa Cruz Biotechnology, sc-502, RRID:AB_631523), and CBP (Santa Cruz Biotechnology, sc-7300, RRID:AB_626817) antibodies were used for WB detection. In figure 1A and supplementary figure S2A, the same blot was probed with cyclin D1 and tubulin antibodies by cutting the membrane. In figure 2H, cyclin D1 and CBP blots correspond to the same membrane while E2F4 and FOXM1 blots correspond to an independent membrane. Image acquisition was performed with ImageQuant LAS 4000 mini (GE Healthcare). Image processing and quantification were performed with Multi Gauge software (Fujifilm). For qRT-PCR analysis, cDNA was generated from 1 µg RNA with qScript cDNA Synthesis kit (Quantabio). qRT–PCR reaction was performed using SYBR green (Roche). -

Differential Expression Profile Analysis of DNA Damage Repair Genes in CD133+/CD133‑ Colorectal Cancer Cells
ONCOLOGY LETTERS 14: 2359-2368, 2017 Differential expression profile analysis of DNA damage repair genes in CD133+/CD133‑ colorectal cancer cells YUHONG LU1*, XIN ZHOU2*, QINGLIANG ZENG2, DAISHUN LIU3 and CHANGWU YUE3 1College of Basic Medicine, Zunyi Medical University, Zunyi; 2Deparment of Gastroenterological Surgery, Affiliated Hospital of Zunyi Medical University, Zunyi;3 Zunyi Key Laboratory of Genetic Diagnosis and Targeted Drug Therapy, The First People's Hospital of Zunyi, Zunyi, Guizhou 563003, P.R. China Received July 20, 2015; Accepted January 6, 2017 DOI: 10.3892/ol.2017.6415 Abstract. The present study examined differential expression cells. By contrast, 6 genes were downregulated and none levels of DNA damage repair genes in COLO 205 colorectal were upregulated in the CD133+ cells compared with the cancer cells, with the aim of identifying novel biomarkers for COLO 205 cells. These findings suggest that CD133+ cells the molecular diagnosis and treatment of colorectal cancer. may possess the same DNA repair capacity as COLO 205 COLO 205-derived cell spheres were cultured in serum-free cells. Heterogeneity in the expression profile of DNA damage medium supplemented with cell factors, and CD133+/CD133- repair genes was observed in COLO 205 cells, and COLO cells were subsequently sorted using an indirect CD133 205-derived CD133- cells and CD133+ cells may therefore microbead kit. In vitro differentiation and tumorigenicity assays provide a reference for molecular diagnosis, therapeutic target in BABA/c nude mice were performed to determine whether selection and determination of the treatment and prognosis for the CD133+ cells also possessed stem cell characteristics, in colorectal cancer. -
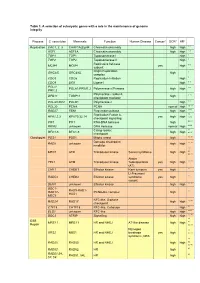
Table 1. a Selection of Eukaryotic Genes with a Role in the Maintenance of Genome Integrity
Table 1. A selection of eukaryotic genes with a role in the maintenance of genome integrity Process S. cerevisiae Mammals Function Human Disease Cancer* GCR* HR* Replication CAC1, 2, 3 CHAF1A,B,p48 Chromatin assembly high high 1, 2 1, 2 ASF1 ASF1A Chromatin assembly high high 3 TOP1 TOP1 Topoisomerase I high 3 TOP2 TOP2 Topoisomerase II high Replicative helicase 4, 5 MCM4 MCM4 yes high subunit Origin Replication 6 ORC3/5 ORC3/6L high complex 7 CDC6 CDC6 Replication Initiation high 7, 8 CDC9 LIG1 Ligase I high POL1/ 7-10 POLA1/PRIM1,2 Polymerase α/Primase high high PRI1,2 Polymerase ε subunit, 6, 11 DPB11 TOBP11 high checkpoint mediator POL3/CDC2 POLD1 Polymerase δ high 7, 8 POL30 PCNA PCNA normal high 12, 13 RAD27 FEN1 Flap endonuclease high high 14-16 Replication Factor A, 14, RFA1,2,3 RPA70,32,14 yes high high 17-19 checkpoint signalling PIF1 PIF1 RNA-DNA helicase high 20, 21 RRM3 unknown DNA Helicase normal high 22-26 Clamp loader, 11, RFC1-5 RFC1-5 high high 20, 27 checkpoint Checkpoint PDS1 PDS1 Mitotic arrest high 11, 28 Damage checkpoint 11, 29 RAD9 unknown high high mediator 11, MEC1 ATR Transducer kinase Seckel syndrome high high 28, 30, 31 Ataxia TEL1 ATM Transducer kinase Telangiectasia yes high high 11, 28 (AT) CHK1 CHEK1 Effector kinase Rare tumours yes high 11 Li-Fraumeni RAD53 CHEK2 Effector kinase syndrome yes high 11 variant DUN1 unknown Effector kinase high high 11, 32 DDC1- RAD9-RAD1- 11 RAD17- PCNA-like complex high HUS1 MEC3 RFC-like, S-phase 11, 33 RAD24 RAD17 high high checkponit 34 CTF18 CHTF18 RFC-like, Cohesion -

The Genetic Program of Pancreatic Beta-Cell Replication in Vivo
Page 1 of 65 Diabetes The genetic program of pancreatic beta-cell replication in vivo Agnes Klochendler1, Inbal Caspi2, Noa Corem1, Maya Moran3, Oriel Friedlich1, Sharona Elgavish4, Yuval Nevo4, Aharon Helman1, Benjamin Glaser5, Amir Eden3, Shalev Itzkovitz2, Yuval Dor1,* 1Department of Developmental Biology and Cancer Research, The Institute for Medical Research Israel-Canada, The Hebrew University-Hadassah Medical School, Jerusalem 91120, Israel 2Department of Molecular Cell Biology, Weizmann Institute of Science, Rehovot, Israel. 3Department of Cell and Developmental Biology, The Silberman Institute of Life Sciences, The Hebrew University of Jerusalem, Jerusalem 91904, Israel 4Info-CORE, Bioinformatics Unit of the I-CORE Computation Center, The Hebrew University and Hadassah, The Institute for Medical Research Israel- Canada, The Hebrew University-Hadassah Medical School, Jerusalem 91120, Israel 5Endocrinology and Metabolism Service, Department of Internal Medicine, Hadassah-Hebrew University Medical Center, Jerusalem 91120, Israel *Correspondence: [email protected] Running title: The genetic program of pancreatic β-cell replication 1 Diabetes Publish Ahead of Print, published online March 18, 2016 Diabetes Page 2 of 65 Abstract The molecular program underlying infrequent replication of pancreatic beta- cells remains largely inaccessible. Using transgenic mice expressing GFP in cycling cells we sorted live, replicating beta-cells and determined their transcriptome. Replicating beta-cells upregulate hundreds of proliferation- related genes, along with many novel putative cell cycle components. Strikingly, genes involved in beta-cell functions, namely glucose sensing and insulin secretion were repressed. Further studies using single molecule RNA in situ hybridization revealed that in fact, replicating beta-cells double the amount of RNA for most genes, but this upregulation excludes genes involved in beta-cell function. -

CHAF1A–PCNA Interaction Promotes Cervical Cancer Progression Via the PI3K/AKT/Foxo1 Signaling Pathway
CHAF1A–PCNA interaction promotes cervical cancer progression via the PI3K/AKT/FoxO1 signaling pathway Li Wen Department of Gynecology, the First Aliated Hospital of Chongqing Medical University Yuli Luo Obstetrics and Gynecology, People's Hospital of Hechuan District, Chongqing Municipality Zhaoning Duan Department of Gynecology, the First Aliated Hospital of Chongqing Medical University Xiaoge Li Department of Gynecology, the First Aliated Hospital of Chongqing Medical University Ying Jia ( [email protected] ) Department of Gynecology, the First Aliated Hospital of Chongqing Medical University https://orcid.org/0000-0002-1602-3122 Primary research Keywords: Cervical cancerCHAF1APCNAInteraction Posted Date: April 12th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-390228/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/18 Abstract Background An increasing number of studies demonstrate that histone chaperones play critical roles in tumorigenesis and development. In previous research, we conrmed that CHAF1A was highly expressed in cervical cancer (CC) and was correlated with poor prognosis. However, the biological function and specic mechanism of CHAF1A in the development of CC have not been reported. Methods CHAF1A knockdown in SiHa and HeLa.cells by lentivirus vector is veried by RT-PCR and Western blot analysis.CCK-8, ow cytometry assays, colony formation, cell migration assay and real-time cell analysis assay were performed to determine the cellular function of CHAF1A in CC.Tumor xenograft assay was conducted on nude mice to assess the effect of CHAF1A in vivo. Cell immunouorescence and co- immunoprecipitation were applied to examine the interaction between CHAF1A and PCNA. -

The HUSH Complex Cooperates with TRIM28 to Repress Young Retrotransposons and New Genes
Downloaded from genome.cshlp.org on September 24, 2021 - Published by Cold Spring Harbor Laboratory Press Research The HUSH complex cooperates with TRIM28 to repress young retrotransposons and new genes Luisa Robbez-Masson,1 Christopher H.C. Tie,1 Lucia Conde,2 Hale Tunbak,1 Connor Husovsky,1 Iva A. Tchasovnikarova,3 Richard T. Timms,3 Javier Herrero,2 Paul J. Lehner,3 and Helen M. Rowe1 1Infection and Immunity, University College London, London WC1E 6BT, United Kingdom; 2Bill Lyons Informatics Centre, UCL Cancer Institute, University College London, London WC1E 6DD, United Kingdom; 3Cambridge Institute for Medical Research, University of Cambridge, Cambridge CB2 0XY, United Kingdom Retrotransposons encompass half of the human genome and contribute to the formation of heterochromatin, which pro- vides nuclear structure and regulates gene expression. Here, we asked if the human silencing hub (HUSH) complex is nec- essary to silence retrotransposons and whether it collaborates with TRIM28 and the chromatin remodeler ATRX at specific genomic loci. We show that the HUSH complex contributes to de novo repression and DNA methylation of an SVA retro- transposon reporter. By using naıvë versus primed mouse pluripotent stem cells, we reveal a critical role for the HUSH com- plex in naıvë cells, implicating it in programming epigenetic marks in development. Although the HUSH component FAM208A binds to endogenous retroviruses (ERVs) and long interspersed element-1s (LINE-1s or L1s), it is mainly required to repress evolutionarily young L1s (mouse-specific lineages <5 million years old). TRIM28, in contrast, is necessary to repress both ERVs and young L1s. Genes co-repressed by TRIM28 and FAM208A are evolutionarily young, or exhibit tissue-specific expression, are enriched in young L1s, and display evidence for regulation through LTR promoters. -

Cell Cycle Arrest Through Indirect Transcriptional Repression by P53: I Have a DREAM
Cell Death and Differentiation (2018) 25, 114–132 Official journal of the Cell Death Differentiation Association OPEN www.nature.com/cdd Review Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM Kurt Engeland1 Activation of the p53 tumor suppressor can lead to cell cycle arrest. The key mechanism of p53-mediated arrest is transcriptional downregulation of many cell cycle genes. In recent years it has become evident that p53-dependent repression is controlled by the p53–p21–DREAM–E2F/CHR pathway (p53–DREAM pathway). DREAM is a transcriptional repressor that binds to E2F or CHR promoter sites. Gene regulation and deregulation by DREAM shares many mechanistic characteristics with the retinoblastoma pRB tumor suppressor that acts through E2F elements. However, because of its binding to E2F and CHR elements, DREAM regulates a larger set of target genes leading to regulatory functions distinct from pRB/E2F. The p53–DREAM pathway controls more than 250 mostly cell cycle-associated genes. The functional spectrum of these pathway targets spans from the G1 phase to the end of mitosis. Consequently, through downregulating the expression of gene products which are essential for progression through the cell cycle, the p53–DREAM pathway participates in the control of all checkpoints from DNA synthesis to cytokinesis including G1/S, G2/M and spindle assembly checkpoints. Therefore, defects in the p53–DREAM pathway contribute to a general loss of checkpoint control. Furthermore, deregulation of DREAM target genes promotes chromosomal instability and aneuploidy of cancer cells. Also, DREAM regulation is abrogated by the human papilloma virus HPV E7 protein linking the p53–DREAM pathway to carcinogenesis by HPV.Another feature of the pathway is that it downregulates many genes involved in DNA repair and telomere maintenance as well as Fanconi anemia. -

SUPPORTING INFORMATION for Regulation of Gene Expression By
SUPPORTING INFORMATION for Regulation of gene expression by the BLM helicase correlates with the presence of G4 motifs Giang Huong Nguyen1,2, Weiliang Tang3, Ana I. Robles1, Richard P. Beyer4, Lucas T. Gray5, Judith A. Welsh1, Aaron J. Schetter1, Kensuke Kumamoto1,6, Xin Wei Wang1, Ian D. Hickson2,7, Nancy Maizels5, 3,8 1 Raymond J. Monnat, Jr. and Curtis C. Harris 1Laboratory of Human Carcinogenesis, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, U.S.A; 2Department of Medical Oncology, Weatherall Institute of Molecular Medicine, John Radcliffe Hospital, University of Oxford, Oxford, U.K.; 3Department of Pathology, University of Washington, Seattle, WA U.S.A.; 4 Center for Ecogenetics and Environmental Health, University of Washington, Seattle, WA U.S.A.; 5Department of Immunology and Department of Biochemistry, University of Washington, Seattle, WA U.S.A.; 6Department of Organ Regulatory Surgery, Fukushima Medical University, Fukushima, Japan; 7Cellular and Molecular Medicine, Nordea Center for Healthy Aging, University of Copenhagen, Denmark; 8Department of Genome Sciences, University of WA, Seattle, WA U.S.A. SI Index: Supporting Information for this manuscript includes the following 19 items. A more detailed Materials and Methods section is followed by 18 Tables and Figures in order of their appearance in the manuscript text: 1) SI Materials and Methods 2) Figure S1. Study design and experimental workflow. 3) Figure S2. Immunoblot verification of BLM depletion from human fibroblasts. 4) Figure S3. PCA of mRNA and miRNA expression in BLM-depleted human fibroblasts. 5) Figure S4. qPCR confirmation of mRNA array data. 6) Table S1. BS patient and control detail. -

CHAF1A (D77D5) XP Rabbit Mab A
Revision 1 C 0 2 ® - t CHAF1A (D77D5) XP Rabbit mAb a e r o t S Orders: 877-616-CELL (2355) [email protected] Support: 877-678-TECH (8324) 0 8 Web: [email protected] 4 www.cellsignal.com 5 # 3 Trask Lane Danvers Massachusetts 01923 USA For Research Use Only. Not For Use In Diagnostic Procedures. Applications: Reactivity: Sensitivity: MW (kDa): Source/Isotype: UniProt ID: Entrez-Gene Id: WB, IP, IF-IC H Mk Endogenous 145 Rabbit IgG Q13111 10036 Product Usage Information 7. Polo, S.E. et al. (2004) Cancer Res 64, 2371-81. Application Dilution Western Blotting 1:1000 Immunoprecipitation 1:100 Immunofluorescence (Immunocytochemistry) 1:200 Storage Supplied in 10 mM sodium HEPES (pH 7.5), 150 mM NaCl, 100 µg/ml BSA, 50% glycerol and less than 0.02% sodium azide. Store at –20°C. Do not aliquot the antibody. Specificity / Sensitivity CHAF1A (D77D5) XP® Rabbit mAb detects endogenous levels of total CHAF1A protein. Species Reactivity: Human, Monkey Source / Purification Monoclonal antibody is produced by immunizing animals with a synthetic peptide corresponding to residues surrounding Pro303 of human CHAF1A protein. Background Chromatin assembly factor 1 (CAF-1) is a histone H3/H4 chaperone complex that functions in de novo assembly of nucleosomes during DNA replication and nucleotide excision repair (1). Nucleosome assembly is a two-step process, involving initial deposition of a histone H3/H4 tetramer onto DNA, followed by the deposition of a pair of histone H2A/H2B dimers (1). CAF-1 interacts with PCNA and localizes to DNA replication and DNA repair foci, where it functions to assemble newly synthesized histone H3/H4 tetramers onto replicating DNA (2-6). -

Host Protein Interactions
scientificscientificreport report Structure homology and interaction redundancy for discovering virus–host protein interactions Benoıˆt de Chassey 1,Laure`ne Meyniel-Schicklin 2,3, Anne Aublin-Gex 2,3, Vincent Navratil 2,3,w, Thibaut Chantier 2,3, Patrice Andre´1,2,3 &VincentLotteau2,3+ 1Hospices Civils de Lyon, Hoˆpital de la Croix-Rousse, Laboratoire de virologie, Lyon , 2Inserm, U1111, Lyon , and 3Universite´ de Lyon, Lyon, France Virus–host interactomes are instrumental to understand global purification coupled to mass spectrometry, led to the publication perturbations of cellular functions induced by infection and of the first virus–host interactomes [3–4]. Although incorporation discover new therapies. The construction of such interactomes is, of data from different methods or variation of the same method [5] however, technically challenging and time consuming. Here we has improved the quality of the data sets, diversification of describe an original method for the prediction of high-confidence methods is still clearly needed to generate high-quality interactions between viral and human proteins through comprehensive virus–host interactomes. In addition, regarding a combination of structure and high-quality interactome data. the size of host genome and the huge diversity of viruses, millions Validation was performed for the NS1 protein of the influenza of binary interactions remain to be tested. Therefore, accurate and virus, which led to the identification of new host factors that rapid methods for the identification of cellular interactors control viral replication. controlling viral replication is a major issue and a crucial step Keywords: interactome; prediction; protein interaction; towards the selection of original therapeutic targets and drug structure; virus development. -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology published online 1 July 2016 from submission to initial decision 4 weeks from acceptance to publication http://www.jimmunol.org/content/early/2016/07/01/jimmun ol.1600589 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol Submit online. Every submission reviewed by practicing scientists ? is published twice each month by http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. Published July 1, 2016, doi:10.4049/jimmunol.1600589 The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh,* Sonia M. Leach,† Brandon L. Moore,* Tullia C. Bruno,* Jonathan D. Buhrman,* and Jill E.