Regulation of Oxidized Base Damage Repair by Chromatin Assembly Factor 1 Subunit a Chunying Yang1,*,†, Shiladitya Sengupta1,2,*,†, Pavana M
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Data
SUPPLEMENTARY DATA A cyclin D1-dependent transcriptional program predicts clinical outcome in mantle cell lymphoma Santiago Demajo et al. 1 SUPPLEMENTARY DATA INDEX Supplementary Methods p. 3 Supplementary References p. 8 Supplementary Tables (S1 to S5) p. 9 Supplementary Figures (S1 to S15) p. 17 2 SUPPLEMENTARY METHODS Western blot, immunoprecipitation, and qRT-PCR Western blot (WB) analysis was performed as previously described (1), using cyclin D1 (Santa Cruz Biotechnology, sc-753, RRID:AB_2070433) and tubulin (Sigma-Aldrich, T5168, RRID:AB_477579) antibodies. Co-immunoprecipitation assays were performed as described before (2), using cyclin D1 antibody (Santa Cruz Biotechnology, sc-8396, RRID:AB_627344) or control IgG (Santa Cruz Biotechnology, sc-2025, RRID:AB_737182) followed by protein G- magnetic beads (Invitrogen) incubation and elution with Glycine 100mM pH=2.5. Co-IP experiments were performed within five weeks after cell thawing. Cyclin D1 (Santa Cruz Biotechnology, sc-753), E2F4 (Bethyl, A302-134A, RRID:AB_1720353), FOXM1 (Santa Cruz Biotechnology, sc-502, RRID:AB_631523), and CBP (Santa Cruz Biotechnology, sc-7300, RRID:AB_626817) antibodies were used for WB detection. In figure 1A and supplementary figure S2A, the same blot was probed with cyclin D1 and tubulin antibodies by cutting the membrane. In figure 2H, cyclin D1 and CBP blots correspond to the same membrane while E2F4 and FOXM1 blots correspond to an independent membrane. Image acquisition was performed with ImageQuant LAS 4000 mini (GE Healthcare). Image processing and quantification were performed with Multi Gauge software (Fujifilm). For qRT-PCR analysis, cDNA was generated from 1 µg RNA with qScript cDNA Synthesis kit (Quantabio). qRT–PCR reaction was performed using SYBR green (Roche). -

Differential Requirements for Tousled-Like Kinases 1 and 2 in Mammalian Development
Cell Death and Differentiation (2017) 24, 1872–1885 & 2017 Macmillan Publishers Limited, part of Springer Nature. All rights reserved 1350-9047/17 www.nature.com/cdd Differential requirements for Tousled-like kinases 1 and 2 in mammalian development Sandra Segura-Bayona1,8, Philip A Knobel1,8, Helena González-Burón1,8, Sameh A Youssef2,3, Aida Peña-Blanco1, Étienne Coyaud4,5, Teresa López-Rovira1, Katrin Rein1, Lluís Palenzuela1, Julien Colombelli1, Stephen Forrow1, Brian Raught4,5, Anja Groth6, Alain de Bruin2,7 and Travis H Stracker*,1 The regulation of chromatin structure is critical for a wide range of essential cellular processes. The Tousled-like kinases, TLK1 and TLK2, regulate ASF1, a histone H3/H4 chaperone, and likely other substrates, and their activity has been implicated in transcription, DNA replication, DNA repair, RNA interference, cell cycle progression, viral latency, chromosome segregation and mitosis. However, little is known about the functions of TLK activity in vivo or the relative functions of the highly similar TLK1 and TLK2 in any cell type. To begin to address this, we have generated Tlk1- and Tlk2-deficient mice. We found that while TLK1 was dispensable for murine viability, TLK2 loss led to late embryonic lethality because of placental failure. TLK2 was required for normal trophoblast differentiation and the phosphorylation of ASF1 was reduced in placentas lacking TLK2. Conditional bypass of the placental phenotype allowed the generation of apparently healthy Tlk2-deficient mice, while only the depletion of both TLK1 and TLK2 led to extensive genomic instability, indicating that both activities contribute to genome maintenance. Our data identifies a specific role for TLK2 in placental function during mammalian development and suggests that TLK1 and TLK2 have largely redundant roles in genome maintenance. -

Histone Chaperone CHAF1A Inhibits Differentiation and Promotes Aggressive Neuroblastoma
Published OnlineFirst December 12, 2013; DOI: 10.1158/0008-5472.CAN-13-1315 Cancer Molecular and Cellular Pathobiology Research Histone Chaperone CHAF1A Inhibits Differentiation and Promotes Aggressive Neuroblastoma Eveline Barbieri1, Katleen De Preter3, Mario Capasso4, Zaowen Chen1, Danielle M. Hsu2, Gian Paolo Tonini5, Steve Lefever3, John Hicks1, Rogier Versteeg7, Andrea Pession6, Frank Speleman3, Eugene S. Kim2, and Jason M. Shohet1 Abstract Neuroblastoma arises from the embryonal neural crest secondary to a block in differentiation. Long-term patient survival correlates inversely with the extent of differentiation, and treatment with retinoic acid or other prodifferentiation agents improves survival modestly. In this study, we show the histone chaperone and epigenetic regulator CHAF1A functions in maintaining the highly dedifferentiated state of this aggressive malignancy. CHAF1A is a subunit of the chromatin modifier chromatin assembly factor 1 and it regulates H3K9 trimethylation of key target genes regulating proliferation, survival, and differentiation. Elevated CHAF1A expression strongly correlated with poor prognosis. Conversely, CHAF1A loss-of-function was sufficient to drive neuronal differentiation in vitro and in vivo. Transcriptome analysis of cells lacking CHAF1A revealed repression of oncogenic signaling pathways and a normalization of glycolytic metabolism. Our findings demonstrate that CHAF1A restricts neural crest differentiation and contributes to the pathogenesis of high-risk neuroblastoma. Cancer Res; 74(3); 1–10. -

TLK2 Antibody (Center) Purified Rabbit Polyclonal Antibody (Pab) Catalog # Ap8102c
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 TLK2 Antibody (Center) Purified Rabbit Polyclonal Antibody (Pab) Catalog # AP8102c Specification TLK2 Antibody (Center) - Product Information Application WB,E Primary Accession Q86UE8 Other Accession Q9UKI7 Reactivity Human, Mouse Host Rabbit Clonality Polyclonal Isotype Rabbit Ig Antigen Region 141-171 TLK2 Antibody (Center) - Additional Information Western blot analysis of anti-TLK2 Antibody Gene ID 11011 (Center) (Cat.#AP8102c) in mouse testis tissue lysates (35ug/lane).TLK2(arrow) was Other Names detected using the purified Pab. Serine/threonine-protein kinase tousled-like 2, HsHPK, PKU-alpha, Tousled-like kinase 2, TLK2 Target/Specificity This TLK2 antibody is generated from rabbits immunized with a KLH conjugated synthetic peptide between 141-171 amino acids from the Central region of human TLK2. Dilution WB~~1:1000 Format Purified polyclonal antibody supplied in PBS TLK2 Antibody (K155) (Cat. #AP8102c) with 0.09% (W/V) sodium azide. This western blot analysis in Y79 cell line lysates antibody is prepared by Saturated (35ug/lane).This demonstrates the TLK2 Ammonium Sulfate (SAS) precipitation antibody detected the TLK2 protein (arrow). followed by dialysis against PBS. Storage TLK2 Antibody (Center) - Background Maintain refrigerated at 2-8°C for up to 2 weeks. For long term storage store at -20°C in small aliquots to prevent freeze-thaw TLK2, a member of the Ser/Thr protein kinase cycles. family, is rapidly and transiently inhibited by phosphorylation following the generation of Precautions DNA double-stranded breaks during S-phase. TLK2 Antibody (Center) is for research use This is cell cycle checkpoint and ATM-pathway only and not for use in diagnostic or dependent and appears to regulate processes therapeutic procedures. -

Differential Expression Profile Analysis of DNA Damage Repair Genes in CD133+/CD133‑ Colorectal Cancer Cells
ONCOLOGY LETTERS 14: 2359-2368, 2017 Differential expression profile analysis of DNA damage repair genes in CD133+/CD133‑ colorectal cancer cells YUHONG LU1*, XIN ZHOU2*, QINGLIANG ZENG2, DAISHUN LIU3 and CHANGWU YUE3 1College of Basic Medicine, Zunyi Medical University, Zunyi; 2Deparment of Gastroenterological Surgery, Affiliated Hospital of Zunyi Medical University, Zunyi;3 Zunyi Key Laboratory of Genetic Diagnosis and Targeted Drug Therapy, The First People's Hospital of Zunyi, Zunyi, Guizhou 563003, P.R. China Received July 20, 2015; Accepted January 6, 2017 DOI: 10.3892/ol.2017.6415 Abstract. The present study examined differential expression cells. By contrast, 6 genes were downregulated and none levels of DNA damage repair genes in COLO 205 colorectal were upregulated in the CD133+ cells compared with the cancer cells, with the aim of identifying novel biomarkers for COLO 205 cells. These findings suggest that CD133+ cells the molecular diagnosis and treatment of colorectal cancer. may possess the same DNA repair capacity as COLO 205 COLO 205-derived cell spheres were cultured in serum-free cells. Heterogeneity in the expression profile of DNA damage medium supplemented with cell factors, and CD133+/CD133- repair genes was observed in COLO 205 cells, and COLO cells were subsequently sorted using an indirect CD133 205-derived CD133- cells and CD133+ cells may therefore microbead kit. In vitro differentiation and tumorigenicity assays provide a reference for molecular diagnosis, therapeutic target in BABA/c nude mice were performed to determine whether selection and determination of the treatment and prognosis for the CD133+ cells also possessed stem cell characteristics, in colorectal cancer. -

Induce Latent/Quiescent HSV-1 Genomes
Promyelocytic leukemia (PML) nuclear bodies (NBs) induce latent/quiescent HSV-1 genomes chromatinization through a PML NB/Histone H3.3/H3.3 Chaperone Axis Camille Cohen, Armelle Corpet, Simon Roubille, Mohamed Maroui, Nolwenn Poccardi, Antoine Rousseau, Constance Kleijwegt, Olivier Binda, Pascale Texier, Nancy Sawtell, et al. To cite this version: Camille Cohen, Armelle Corpet, Simon Roubille, Mohamed Maroui, Nolwenn Poccardi, et al.. Promye- locytic leukemia (PML) nuclear bodies (NBs) induce latent/quiescent HSV-1 genomes chromatiniza- tion through a PML NB/Histone H3.3/H3.3 Chaperone Axis. PLoS Pathogens, Public Library of Science, 2018, 14 (9), pp.e1007313. 10.1371/journal.ppat.1007313. inserm-02167220 HAL Id: inserm-02167220 https://www.hal.inserm.fr/inserm-02167220 Submitted on 27 Jun 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. RESEARCH ARTICLE Promyelocytic leukemia (PML) nuclear bodies (NBs) induce latent/quiescent HSV-1 genomes chromatinization through a PML NB/Histone H3.3/H3.3 Chaperone Axis Camille Cohen1, Armelle Corpet1, Simon -

Histone H3.3 and Its Proteolytically Processed Form Drive a Cellular Senescence Programme
ARTICLE Received 21 Mar 2014 | Accepted 9 Sep 2014 | Published 14 Nov 2014 DOI: 10.1038/ncomms6210 Histone H3.3 and its proteolytically processed form drive a cellular senescence programme Luis F. Duarte1,2,3, Andrew R.J. Young4, Zichen Wang3,5, Hsan-Au Wu1,3, Taniya Panda1,2, Yan Kou3,5, Avnish Kapoor1,2,w, Dan Hasson1,2, Nicholas R. Mills1,2, Avi Ma’ayan5, Masashi Narita4 & Emily Bernstein1,2 The process of cellular senescence generates a repressive chromatin environment, however, the role of histone variants and histone proteolytic cleavage in senescence remains unclear. Here, using models of oncogene-induced and replicative senescence, we report novel histone H3 tail cleavage events mediated by the protease Cathepsin L. We find that cleaved forms of H3 are nucleosomal and the histone variant H3.3 is the preferred cleaved form of H3. Ectopic expression of H3.3 and its cleavage product (H3.3cs1), which lacks the first 21 amino acids of the H3 tail, is sufficient to induce senescence. Further, H3.3cs1 chromatin incorporation is mediated by the HUCA histone chaperone complex. Genome-wide transcriptional profiling revealed that H3.3cs1 facilitates transcriptional silencing of cell cycle regulators including RB/E2F target genes, likely via the permanent removal of H3K4me3. Collectively, our study identifies histone H3.3 and its proteolytically processed forms as key regulators of cellular senescence. 1 Department of Oncological Sciences, Icahn School of Medicine at Mount Sinai, One Gustave L. Levy Place, New York, New York 10029, USA. 2 Department of Dermatology, Icahn School of Medicine at Mount Sinai, One Gustave L. -

Research2007herschkowitzetvolume Al
Open Access Research2007HerschkowitzetVolume al. 8, Issue 5, Article R76 Identification of conserved gene expression features between comment murine mammary carcinoma models and human breast tumors Jason I Herschkowitz¤*†, Karl Simin¤‡, Victor J Weigman§, Igor Mikaelian¶, Jerry Usary*¥, Zhiyuan Hu*¥, Karen E Rasmussen*¥, Laundette P Jones#, Shahin Assefnia#, Subhashini Chandrasekharan¥, Michael G Backlund†, Yuzhi Yin#, Andrey I Khramtsov**, Roy Bastein††, John Quackenbush††, Robert I Glazer#, Powel H Brown‡‡, Jeffrey E Green§§, Levy Kopelovich, reviews Priscilla A Furth#, Juan P Palazzo, Olufunmilayo I Olopade, Philip S Bernard††, Gary A Churchill¶, Terry Van Dyke*¥ and Charles M Perou*¥ Addresses: *Lineberger Comprehensive Cancer Center. †Curriculum in Genetics and Molecular Biology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA. ‡Department of Cancer Biology, University of Massachusetts Medical School, Worcester, MA 01605, USA. reports §Department of Biology and Program in Bioinformatics and Computational Biology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA. ¶The Jackson Laboratory, Bar Harbor, ME 04609, USA. ¥Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA. #Department of Oncology, Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC 20057, USA. **Department of Pathology, University of Chicago, Chicago, IL 60637, USA. ††Department of Pathology, University of Utah School of Medicine, Salt Lake City, UT 84132, USA. ‡‡Baylor College of Medicine, Houston, TX 77030, USA. §§Transgenic Oncogenesis Group, Laboratory of Cancer Biology and Genetics. Chemoprevention Agent Development Research Group, National Cancer Institute, Bethesda, MD 20892, USA. Department of Pathology, Thomas Jefferson University, Philadelphia, PA 19107, USA. Section of Hematology/Oncology, Department of Medicine, Committees on Genetics and Cancer Biology, University of Chicago, Chicago, IL 60637, USA. -

A High-Throughput Approach to Uncover Novel Roles of APOBEC2, a Functional Orphan of the AID/APOBEC Family
Rockefeller University Digital Commons @ RU Student Theses and Dissertations 2018 A High-Throughput Approach to Uncover Novel Roles of APOBEC2, a Functional Orphan of the AID/APOBEC Family Linda Molla Follow this and additional works at: https://digitalcommons.rockefeller.edu/ student_theses_and_dissertations Part of the Life Sciences Commons A HIGH-THROUGHPUT APPROACH TO UNCOVER NOVEL ROLES OF APOBEC2, A FUNCTIONAL ORPHAN OF THE AID/APOBEC FAMILY A Thesis Presented to the Faculty of The Rockefeller University in Partial Fulfillment of the Requirements for the degree of Doctor of Philosophy by Linda Molla June 2018 © Copyright by Linda Molla 2018 A HIGH-THROUGHPUT APPROACH TO UNCOVER NOVEL ROLES OF APOBEC2, A FUNCTIONAL ORPHAN OF THE AID/APOBEC FAMILY Linda Molla, Ph.D. The Rockefeller University 2018 APOBEC2 is a member of the AID/APOBEC cytidine deaminase family of proteins. Unlike most of AID/APOBEC, however, APOBEC2’s function remains elusive. Previous research has implicated APOBEC2 in diverse organisms and cellular processes such as muscle biology (in Mus musculus), regeneration (in Danio rerio), and development (in Xenopus laevis). APOBEC2 has also been implicated in cancer. However the enzymatic activity, substrate or physiological target(s) of APOBEC2 are unknown. For this thesis, I have combined Next Generation Sequencing (NGS) techniques with state-of-the-art molecular biology to determine the physiological targets of APOBEC2. Using a cell culture muscle differentiation system, and RNA sequencing (RNA-Seq) by polyA capture, I demonstrated that unlike the AID/APOBEC family member APOBEC1, APOBEC2 is not an RNA editor. Using the same system combined with enhanced Reduced Representation Bisulfite Sequencing (eRRBS) analyses I showed that, unlike the AID/APOBEC family member AID, APOBEC2 does not act as a 5-methyl-C deaminase. -
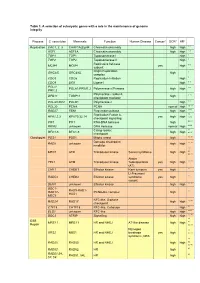
Table 1. a Selection of Eukaryotic Genes with a Role in the Maintenance of Genome Integrity
Table 1. A selection of eukaryotic genes with a role in the maintenance of genome integrity Process S. cerevisiae Mammals Function Human Disease Cancer* GCR* HR* Replication CAC1, 2, 3 CHAF1A,B,p48 Chromatin assembly high high 1, 2 1, 2 ASF1 ASF1A Chromatin assembly high high 3 TOP1 TOP1 Topoisomerase I high 3 TOP2 TOP2 Topoisomerase II high Replicative helicase 4, 5 MCM4 MCM4 yes high subunit Origin Replication 6 ORC3/5 ORC3/6L high complex 7 CDC6 CDC6 Replication Initiation high 7, 8 CDC9 LIG1 Ligase I high POL1/ 7-10 POLA1/PRIM1,2 Polymerase α/Primase high high PRI1,2 Polymerase ε subunit, 6, 11 DPB11 TOBP11 high checkpoint mediator POL3/CDC2 POLD1 Polymerase δ high 7, 8 POL30 PCNA PCNA normal high 12, 13 RAD27 FEN1 Flap endonuclease high high 14-16 Replication Factor A, 14, RFA1,2,3 RPA70,32,14 yes high high 17-19 checkpoint signalling PIF1 PIF1 RNA-DNA helicase high 20, 21 RRM3 unknown DNA Helicase normal high 22-26 Clamp loader, 11, RFC1-5 RFC1-5 high high 20, 27 checkpoint Checkpoint PDS1 PDS1 Mitotic arrest high 11, 28 Damage checkpoint 11, 29 RAD9 unknown high high mediator 11, MEC1 ATR Transducer kinase Seckel syndrome high high 28, 30, 31 Ataxia TEL1 ATM Transducer kinase Telangiectasia yes high high 11, 28 (AT) CHK1 CHEK1 Effector kinase Rare tumours yes high 11 Li-Fraumeni RAD53 CHEK2 Effector kinase syndrome yes high 11 variant DUN1 unknown Effector kinase high high 11, 32 DDC1- RAD9-RAD1- 11 RAD17- PCNA-like complex high HUS1 MEC3 RFC-like, S-phase 11, 33 RAD24 RAD17 high high checkponit 34 CTF18 CHTF18 RFC-like, Cohesion -

The Genetic Program of Pancreatic Beta-Cell Replication in Vivo
Page 1 of 65 Diabetes The genetic program of pancreatic beta-cell replication in vivo Agnes Klochendler1, Inbal Caspi2, Noa Corem1, Maya Moran3, Oriel Friedlich1, Sharona Elgavish4, Yuval Nevo4, Aharon Helman1, Benjamin Glaser5, Amir Eden3, Shalev Itzkovitz2, Yuval Dor1,* 1Department of Developmental Biology and Cancer Research, The Institute for Medical Research Israel-Canada, The Hebrew University-Hadassah Medical School, Jerusalem 91120, Israel 2Department of Molecular Cell Biology, Weizmann Institute of Science, Rehovot, Israel. 3Department of Cell and Developmental Biology, The Silberman Institute of Life Sciences, The Hebrew University of Jerusalem, Jerusalem 91904, Israel 4Info-CORE, Bioinformatics Unit of the I-CORE Computation Center, The Hebrew University and Hadassah, The Institute for Medical Research Israel- Canada, The Hebrew University-Hadassah Medical School, Jerusalem 91120, Israel 5Endocrinology and Metabolism Service, Department of Internal Medicine, Hadassah-Hebrew University Medical Center, Jerusalem 91120, Israel *Correspondence: [email protected] Running title: The genetic program of pancreatic β-cell replication 1 Diabetes Publish Ahead of Print, published online March 18, 2016 Diabetes Page 2 of 65 Abstract The molecular program underlying infrequent replication of pancreatic beta- cells remains largely inaccessible. Using transgenic mice expressing GFP in cycling cells we sorted live, replicating beta-cells and determined their transcriptome. Replicating beta-cells upregulate hundreds of proliferation- related genes, along with many novel putative cell cycle components. Strikingly, genes involved in beta-cell functions, namely glucose sensing and insulin secretion were repressed. Further studies using single molecule RNA in situ hybridization revealed that in fact, replicating beta-cells double the amount of RNA for most genes, but this upregulation excludes genes involved in beta-cell function. -

CHAF1A–PCNA Interaction Promotes Cervical Cancer Progression Via the PI3K/AKT/Foxo1 Signaling Pathway
CHAF1A–PCNA interaction promotes cervical cancer progression via the PI3K/AKT/FoxO1 signaling pathway Li Wen Department of Gynecology, the First Aliated Hospital of Chongqing Medical University Yuli Luo Obstetrics and Gynecology, People's Hospital of Hechuan District, Chongqing Municipality Zhaoning Duan Department of Gynecology, the First Aliated Hospital of Chongqing Medical University Xiaoge Li Department of Gynecology, the First Aliated Hospital of Chongqing Medical University Ying Jia ( [email protected] ) Department of Gynecology, the First Aliated Hospital of Chongqing Medical University https://orcid.org/0000-0002-1602-3122 Primary research Keywords: Cervical cancerCHAF1APCNAInteraction Posted Date: April 12th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-390228/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/18 Abstract Background An increasing number of studies demonstrate that histone chaperones play critical roles in tumorigenesis and development. In previous research, we conrmed that CHAF1A was highly expressed in cervical cancer (CC) and was correlated with poor prognosis. However, the biological function and specic mechanism of CHAF1A in the development of CC have not been reported. Methods CHAF1A knockdown in SiHa and HeLa.cells by lentivirus vector is veried by RT-PCR and Western blot analysis.CCK-8, ow cytometry assays, colony formation, cell migration assay and real-time cell analysis assay were performed to determine the cellular function of CHAF1A in CC.Tumor xenograft assay was conducted on nude mice to assess the effect of CHAF1A in vivo. Cell immunouorescence and co- immunoprecipitation were applied to examine the interaction between CHAF1A and PCNA.