CHAF1A (D77D5) XP Rabbit Mab A
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Regulation of Oxidized Base Damage Repair by Chromatin Assembly Factor 1 Subunit a Chunying Yang1,*,†, Shiladitya Sengupta1,2,*,†, Pavana M
Published online 27 October 2016 Nucleic Acids Research, 2017, Vol. 45, No. 2 739–748 doi: 10.1093/nar/gkw1024 Regulation of oxidized base damage repair by chromatin assembly factor 1 subunit A Chunying Yang1,*,†, Shiladitya Sengupta1,2,*,†, Pavana M. Hegde1,JoyMitra1, Shuai Jiang3, Brooke Holey3, Altaf H. Sarker3, Miaw-Sheue Tsai3, Muralidhar L. Hegde1,2,4 and Sankar Mitra1,2,* 1Department of Radiation Oncology, Houston Methodist Research Institute, Houston, TX 77030, USA, 2Weill Cornell Medical College, Cornell University, New York, NY 10065, USA, 3Biological Systems and Engineering Division, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA and 4Houston Methodist Neurological Institute, Houston, TX 77030, USA Received March 23, 2016; Revised October 13, 2016; Editorial Decision October 17, 2016; Accepted October 19, 2016 ABSTRACT INTRODUCTION Reactive oxygen species (ROS), generated both en- ROS, continuously generated in mammalian cells both en- dogenously and in response to exogenous stress, in- dogenously and by environmental genotoxicants, induce duce point mutations by mis-replication of oxidized various genomic lesions, including oxidized bases, abasic bases and other lesions in the genome. Repair of (AP) sites and single-strand breaks (SSBs). If unrepaired or these lesions via base excision repair (BER) pathway mis-repaired, DNA lesions would cause mutations which may lead to cytotoxicity and cell death and also carcino- maintains genomic fidelity. Regulation of the BER genic transformation (1). The base excision repair (BER) pathway for mutagenic oxidized bases, initiated by pathway, responsible for repair of oxidized base lesions NEIL1 and other DNA glycosylases at the chromatin which contribute to drug/radiation sensitivity is highly con- level remains unexplored. -

Supplementary Data
SUPPLEMENTARY DATA A cyclin D1-dependent transcriptional program predicts clinical outcome in mantle cell lymphoma Santiago Demajo et al. 1 SUPPLEMENTARY DATA INDEX Supplementary Methods p. 3 Supplementary References p. 8 Supplementary Tables (S1 to S5) p. 9 Supplementary Figures (S1 to S15) p. 17 2 SUPPLEMENTARY METHODS Western blot, immunoprecipitation, and qRT-PCR Western blot (WB) analysis was performed as previously described (1), using cyclin D1 (Santa Cruz Biotechnology, sc-753, RRID:AB_2070433) and tubulin (Sigma-Aldrich, T5168, RRID:AB_477579) antibodies. Co-immunoprecipitation assays were performed as described before (2), using cyclin D1 antibody (Santa Cruz Biotechnology, sc-8396, RRID:AB_627344) or control IgG (Santa Cruz Biotechnology, sc-2025, RRID:AB_737182) followed by protein G- magnetic beads (Invitrogen) incubation and elution with Glycine 100mM pH=2.5. Co-IP experiments were performed within five weeks after cell thawing. Cyclin D1 (Santa Cruz Biotechnology, sc-753), E2F4 (Bethyl, A302-134A, RRID:AB_1720353), FOXM1 (Santa Cruz Biotechnology, sc-502, RRID:AB_631523), and CBP (Santa Cruz Biotechnology, sc-7300, RRID:AB_626817) antibodies were used for WB detection. In figure 1A and supplementary figure S2A, the same blot was probed with cyclin D1 and tubulin antibodies by cutting the membrane. In figure 2H, cyclin D1 and CBP blots correspond to the same membrane while E2F4 and FOXM1 blots correspond to an independent membrane. Image acquisition was performed with ImageQuant LAS 4000 mini (GE Healthcare). Image processing and quantification were performed with Multi Gauge software (Fujifilm). For qRT-PCR analysis, cDNA was generated from 1 µg RNA with qScript cDNA Synthesis kit (Quantabio). qRT–PCR reaction was performed using SYBR green (Roche). -

Histone Chaperone CHAF1A Inhibits Differentiation and Promotes Aggressive Neuroblastoma
Published OnlineFirst December 12, 2013; DOI: 10.1158/0008-5472.CAN-13-1315 Cancer Molecular and Cellular Pathobiology Research Histone Chaperone CHAF1A Inhibits Differentiation and Promotes Aggressive Neuroblastoma Eveline Barbieri1, Katleen De Preter3, Mario Capasso4, Zaowen Chen1, Danielle M. Hsu2, Gian Paolo Tonini5, Steve Lefever3, John Hicks1, Rogier Versteeg7, Andrea Pession6, Frank Speleman3, Eugene S. Kim2, and Jason M. Shohet1 Abstract Neuroblastoma arises from the embryonal neural crest secondary to a block in differentiation. Long-term patient survival correlates inversely with the extent of differentiation, and treatment with retinoic acid or other prodifferentiation agents improves survival modestly. In this study, we show the histone chaperone and epigenetic regulator CHAF1A functions in maintaining the highly dedifferentiated state of this aggressive malignancy. CHAF1A is a subunit of the chromatin modifier chromatin assembly factor 1 and it regulates H3K9 trimethylation of key target genes regulating proliferation, survival, and differentiation. Elevated CHAF1A expression strongly correlated with poor prognosis. Conversely, CHAF1A loss-of-function was sufficient to drive neuronal differentiation in vitro and in vivo. Transcriptome analysis of cells lacking CHAF1A revealed repression of oncogenic signaling pathways and a normalization of glycolytic metabolism. Our findings demonstrate that CHAF1A restricts neural crest differentiation and contributes to the pathogenesis of high-risk neuroblastoma. Cancer Res; 74(3); 1–10. -

Differential Expression Profile Analysis of DNA Damage Repair Genes in CD133+/CD133‑ Colorectal Cancer Cells
ONCOLOGY LETTERS 14: 2359-2368, 2017 Differential expression profile analysis of DNA damage repair genes in CD133+/CD133‑ colorectal cancer cells YUHONG LU1*, XIN ZHOU2*, QINGLIANG ZENG2, DAISHUN LIU3 and CHANGWU YUE3 1College of Basic Medicine, Zunyi Medical University, Zunyi; 2Deparment of Gastroenterological Surgery, Affiliated Hospital of Zunyi Medical University, Zunyi;3 Zunyi Key Laboratory of Genetic Diagnosis and Targeted Drug Therapy, The First People's Hospital of Zunyi, Zunyi, Guizhou 563003, P.R. China Received July 20, 2015; Accepted January 6, 2017 DOI: 10.3892/ol.2017.6415 Abstract. The present study examined differential expression cells. By contrast, 6 genes were downregulated and none levels of DNA damage repair genes in COLO 205 colorectal were upregulated in the CD133+ cells compared with the cancer cells, with the aim of identifying novel biomarkers for COLO 205 cells. These findings suggest that CD133+ cells the molecular diagnosis and treatment of colorectal cancer. may possess the same DNA repair capacity as COLO 205 COLO 205-derived cell spheres were cultured in serum-free cells. Heterogeneity in the expression profile of DNA damage medium supplemented with cell factors, and CD133+/CD133- repair genes was observed in COLO 205 cells, and COLO cells were subsequently sorted using an indirect CD133 205-derived CD133- cells and CD133+ cells may therefore microbead kit. In vitro differentiation and tumorigenicity assays provide a reference for molecular diagnosis, therapeutic target in BABA/c nude mice were performed to determine whether selection and determination of the treatment and prognosis for the CD133+ cells also possessed stem cell characteristics, in colorectal cancer. -

Chromosome 21 Leading Edge Gene Set
Chromosome 21 Leading Edge Gene Set Genes from chr21q22 that are part of the GSEA leading edge set identifying differences between trisomic and euploid samples. Multiple probe set IDs corresponding to a single gene symbol are combined as part of the GSEA analysis. Gene Symbol Probe Set IDs Gene Title 203865_s_at, 207999_s_at, 209979_at, adenosine deaminase, RNA-specific, B1 ADARB1 234539_at, 234799_at (RED1 homolog rat) UDP-Gal:betaGlcNAc beta 1,3- B3GALT5 206947_at galactosyltransferase, polypeptide 5 BACE2 217867_x_at, 222446_s_at beta-site APP-cleaving enzyme 2 1553227_s_at, 214820_at, 219280_at, 225446_at, 231860_at, 231960_at, bromodomain and WD repeat domain BRWD1 244622_at containing 1 C21orf121 240809_at chromosome 21 open reading frame 121 C21orf130 240068_at chromosome 21 open reading frame 130 C21orf22 1560881_a_at chromosome 21 open reading frame 22 C21orf29 1552570_at, 1555048_at_at, 1555049_at chromosome 21 open reading frame 29 C21orf33 202217_at, 210667_s_at chromosome 21 open reading frame 33 C21orf45 219004_s_at, 228597_at, 229671_s_at chromosome 21 open reading frame 45 C21orf51 1554430_at, 1554432_x_at, 228239_at chromosome 21 open reading frame 51 C21orf56 223360_at chromosome 21 open reading frame 56 C21orf59 218123_at, 244369_at chromosome 21 open reading frame 59 C21orf66 1555125_at, 218515_at, 221158_at chromosome 21 open reading frame 66 C21orf7 221211_s_at chromosome 21 open reading frame 7 C21orf77 220826_at chromosome 21 open reading frame 77 C21orf84 239968_at, 240589_at chromosome 21 open reading frame 84 -
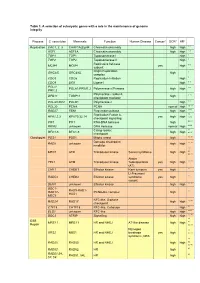
Table 1. a Selection of Eukaryotic Genes with a Role in the Maintenance of Genome Integrity
Table 1. A selection of eukaryotic genes with a role in the maintenance of genome integrity Process S. cerevisiae Mammals Function Human Disease Cancer* GCR* HR* Replication CAC1, 2, 3 CHAF1A,B,p48 Chromatin assembly high high 1, 2 1, 2 ASF1 ASF1A Chromatin assembly high high 3 TOP1 TOP1 Topoisomerase I high 3 TOP2 TOP2 Topoisomerase II high Replicative helicase 4, 5 MCM4 MCM4 yes high subunit Origin Replication 6 ORC3/5 ORC3/6L high complex 7 CDC6 CDC6 Replication Initiation high 7, 8 CDC9 LIG1 Ligase I high POL1/ 7-10 POLA1/PRIM1,2 Polymerase α/Primase high high PRI1,2 Polymerase ε subunit, 6, 11 DPB11 TOBP11 high checkpoint mediator POL3/CDC2 POLD1 Polymerase δ high 7, 8 POL30 PCNA PCNA normal high 12, 13 RAD27 FEN1 Flap endonuclease high high 14-16 Replication Factor A, 14, RFA1,2,3 RPA70,32,14 yes high high 17-19 checkpoint signalling PIF1 PIF1 RNA-DNA helicase high 20, 21 RRM3 unknown DNA Helicase normal high 22-26 Clamp loader, 11, RFC1-5 RFC1-5 high high 20, 27 checkpoint Checkpoint PDS1 PDS1 Mitotic arrest high 11, 28 Damage checkpoint 11, 29 RAD9 unknown high high mediator 11, MEC1 ATR Transducer kinase Seckel syndrome high high 28, 30, 31 Ataxia TEL1 ATM Transducer kinase Telangiectasia yes high high 11, 28 (AT) CHK1 CHEK1 Effector kinase Rare tumours yes high 11 Li-Fraumeni RAD53 CHEK2 Effector kinase syndrome yes high 11 variant DUN1 unknown Effector kinase high high 11, 32 DDC1- RAD9-RAD1- 11 RAD17- PCNA-like complex high HUS1 MEC3 RFC-like, S-phase 11, 33 RAD24 RAD17 high high checkponit 34 CTF18 CHTF18 RFC-like, Cohesion -

The Genetic Program of Pancreatic Beta-Cell Replication in Vivo
Page 1 of 65 Diabetes The genetic program of pancreatic beta-cell replication in vivo Agnes Klochendler1, Inbal Caspi2, Noa Corem1, Maya Moran3, Oriel Friedlich1, Sharona Elgavish4, Yuval Nevo4, Aharon Helman1, Benjamin Glaser5, Amir Eden3, Shalev Itzkovitz2, Yuval Dor1,* 1Department of Developmental Biology and Cancer Research, The Institute for Medical Research Israel-Canada, The Hebrew University-Hadassah Medical School, Jerusalem 91120, Israel 2Department of Molecular Cell Biology, Weizmann Institute of Science, Rehovot, Israel. 3Department of Cell and Developmental Biology, The Silberman Institute of Life Sciences, The Hebrew University of Jerusalem, Jerusalem 91904, Israel 4Info-CORE, Bioinformatics Unit of the I-CORE Computation Center, The Hebrew University and Hadassah, The Institute for Medical Research Israel- Canada, The Hebrew University-Hadassah Medical School, Jerusalem 91120, Israel 5Endocrinology and Metabolism Service, Department of Internal Medicine, Hadassah-Hebrew University Medical Center, Jerusalem 91120, Israel *Correspondence: [email protected] Running title: The genetic program of pancreatic β-cell replication 1 Diabetes Publish Ahead of Print, published online March 18, 2016 Diabetes Page 2 of 65 Abstract The molecular program underlying infrequent replication of pancreatic beta- cells remains largely inaccessible. Using transgenic mice expressing GFP in cycling cells we sorted live, replicating beta-cells and determined their transcriptome. Replicating beta-cells upregulate hundreds of proliferation- related genes, along with many novel putative cell cycle components. Strikingly, genes involved in beta-cell functions, namely glucose sensing and insulin secretion were repressed. Further studies using single molecule RNA in situ hybridization revealed that in fact, replicating beta-cells double the amount of RNA for most genes, but this upregulation excludes genes involved in beta-cell function. -

Increased Dosage of the Chromosome 21 Ortholog Dyrk1a Promotes Megakaryoblastic Leukemia in a Murine Model of Down Syndrome
Increased dosage of the chromosome 21 ortholog Dyrk1a promotes megakaryoblastic leukemia in a murine model of Down syndrome Sébastien Malinge, … , Sandeep Gurbuxani, John D. Crispino J Clin Invest. 2012;122(3):948-962. https://doi.org/10.1172/JCI60455. Research Article Individuals with Down syndrome (DS; also known as trisomy 21) have a markedly increased risk of leukemia in childhood but a decreased risk of solid tumors in adulthood. Acquired mutations in the transcription factor–encoding GATA1 gene are observed in nearly all individuals with DS who are born with transient myeloproliferative disorder (TMD), a clonal preleukemia, and/or who develop acute megakaryoblastic leukemia (AMKL). Individuals who do not have DS but bear germline GATA1 mutations analogous to those detected in individuals with TMD and DS-AMKL are not predisposed to leukemia. To better understand the functional contribution of trisomy 21 to leukemogenesis, we used mouse and human cell models of DS to reproduce the multistep pathogenesis of DS-AMKL and to identify chromosome 21 genes that promote megakaryoblastic leukemia in children with DS. Our results revealed that trisomy for only 33 orthologs of human chromosome 21 (Hsa21) genes was sufficient to cooperate with GATA1 mutations to initiate megakaryoblastic leukemia in vivo. Furthermore, through a functional screening of the trisomic genes, we demonstrated that DYRK1A, which encodes dual-specificity tyrosine-(Y)-phosphorylation–regulated kinase 1A, was a potent megakaryoblastic tumor–promoting gene that contributed -

Network-Based Method for Drug Target Discovery at the Isoform Level
www.nature.com/scientificreports OPEN Network-based method for drug target discovery at the isoform level Received: 20 November 2018 Jun Ma1,2, Jenny Wang2, Laleh Soltan Ghoraie2, Xin Men3, Linna Liu4 & Penggao Dai 1 Accepted: 6 September 2019 Identifcation of primary targets associated with phenotypes can facilitate exploration of the underlying Published: xx xx xxxx molecular mechanisms of compounds and optimization of the structures of promising drugs. However, the literature reports limited efort to identify the target major isoform of a single known target gene. The majority of genes generate multiple transcripts that are translated into proteins that may carry out distinct and even opposing biological functions through alternative splicing. In addition, isoform expression is dynamic and varies depending on the developmental stage and cell type. To identify target major isoforms, we integrated a breast cancer type-specifc isoform coexpression network with gene perturbation signatures in the MCF7 cell line in the Connectivity Map database using the ‘shortest path’ drug target prioritization method. We used a leukemia cancer network and diferential expression data for drugs in the HL-60 cell line to test the robustness of the detection algorithm for target major isoforms. We further analyzed the properties of target major isoforms for each multi-isoform gene using pharmacogenomic datasets, proteomic data and the principal isoforms defned by the APPRIS and STRING datasets. Then, we tested our predictions for the most promising target major protein isoforms of DNMT1, MGEA5 and P4HB4 based on expression data and topological features in the coexpression network. Interestingly, these isoforms are not annotated as principal isoforms in APPRIS. -

CHAF1A–PCNA Interaction Promotes Cervical Cancer Progression Via the PI3K/AKT/Foxo1 Signaling Pathway
CHAF1A–PCNA interaction promotes cervical cancer progression via the PI3K/AKT/FoxO1 signaling pathway Li Wen Department of Gynecology, the First Aliated Hospital of Chongqing Medical University Yuli Luo Obstetrics and Gynecology, People's Hospital of Hechuan District, Chongqing Municipality Zhaoning Duan Department of Gynecology, the First Aliated Hospital of Chongqing Medical University Xiaoge Li Department of Gynecology, the First Aliated Hospital of Chongqing Medical University Ying Jia ( [email protected] ) Department of Gynecology, the First Aliated Hospital of Chongqing Medical University https://orcid.org/0000-0002-1602-3122 Primary research Keywords: Cervical cancerCHAF1APCNAInteraction Posted Date: April 12th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-390228/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/18 Abstract Background An increasing number of studies demonstrate that histone chaperones play critical roles in tumorigenesis and development. In previous research, we conrmed that CHAF1A was highly expressed in cervical cancer (CC) and was correlated with poor prognosis. However, the biological function and specic mechanism of CHAF1A in the development of CC have not been reported. Methods CHAF1A knockdown in SiHa and HeLa.cells by lentivirus vector is veried by RT-PCR and Western blot analysis.CCK-8, ow cytometry assays, colony formation, cell migration assay and real-time cell analysis assay were performed to determine the cellular function of CHAF1A in CC.Tumor xenograft assay was conducted on nude mice to assess the effect of CHAF1A in vivo. Cell immunouorescence and co- immunoprecipitation were applied to examine the interaction between CHAF1A and PCNA. -

Integrated Functional Genomic Analysis Enables Annotation of Kidney Genome-Wide Association Study Loci
BASIC RESEARCH www.jasn.org Integrated Functional Genomic Analysis Enables Annotation of Kidney Genome-Wide Association Study Loci Karsten B. Sieber,1 Anna Batorsky,2 Kyle Siebenthall,2 Kelly L. Hudkins,3 Jeff D. Vierstra,2 Shawn Sullivan,4 Aakash Sur,4,5 Michelle McNulty,6 Richard Sandstrom,2 Alex Reynolds,2 Daniel Bates,2 Morgan Diegel,2 Douglass Dunn,2 Jemma Nelson,2 Michael Buckley,2 Rajinder Kaul,2 Matthew G. Sampson,6 Jonathan Himmelfarb,7,8 Charles E. Alpers,3,8 Dawn Waterworth,1 and Shreeram Akilesh3,8 Due to the number of contributing authors, the affiliations are listed at the end of this article. ABSTRACT Background Linking genetic risk loci identified by genome-wide association studies (GWAS) to their causal genes remains a major challenge. Disease-associated genetic variants are concentrated in regions con- taining regulatory DNA elements, such as promoters and enhancers. Although researchers have previ- ously published DNA maps of these regulatory regions for kidney tubule cells and glomerular endothelial cells, maps for podocytes and mesangial cells have not been available. Methods We generated regulatory DNA maps (DNase-seq) and paired gene expression profiles (RNA-seq) from primary outgrowth cultures of human glomeruli that were composed mainly of podo- cytes and mesangial cells. We generated similar datasets from renal cortex cultures, to compare with those of the glomerular cultures. Because regulatory DNA elements can act on target genes across large genomic distances, we also generated a chromatin conformation map from freshly isolated human glomeruli. Results We identified thousands of unique regulatory DNA elements, many located close to transcription factor genes, which the glomerular and cortex samples expressed at different levels. -

Risk Stratification of Triple-Negative Breast Cancer with Core Gene
Breast Cancer Research and Treatment (2019) 178:185–197 https://doi.org/10.1007/s10549-019-05366-x EPIDEMIOLOGY Risk stratifcation of triple‑negative breast cancer with core gene signatures associated with chemoresponse and prognosis Eun‑Kyu Kim1 · Ae Kyung Park2 · Eunyoung Ko3 · Woong‑Yang Park4 · Kyung‑Min Lee5 · Dong‑Young Noh5,6 · Wonshik Han5,6 Received: 8 May 2019 / Accepted: 16 July 2019 / Published online: 24 July 2019 © Springer Science+Business Media, LLC, part of Springer Nature 2019 Abstract Purpose Neoadjuvant chemotherapy studies have consistently reported a strong correlation between pathologic response and long-term outcome in triple-negative breast cancer (TNBC). We aimed to defne minimal gene signatures for predicting chemoresponse by a three-step approach and to further develop a risk-stratifcation method of TNBC. Methods The frst step involved the detection of genes associated with resistance to docetaxel in eight TNBC cell lines, leading to identifcation of thousands of candidate genes. Through subsequent second and third step analyses with gene set enrichment analysis and survival analysis using public expression profles, the candidate gene list was reduced to prognostic core gene signatures comprising ten or four genes. Results The prognostic core gene signatures include three up-regulated (CEBPD, MMP20, and WLS) and seven down- regulated genes (ASF1A, ASPSCR1, CHAF1B, DNMT1, GINS2, GOLGA2P5, and SKA1). We further develop a simple risk-stratifcation method based on expression profles of the core genes. Relative expression values of the up-regulated and down-regulated core genes were averaged into two scores, Up and Down scores, respectively; then samples were stratifed by a diagonal line in a xy plot of the Up and Down scores.