The Nurr1 Ligand,1,1-Bis(3′-Indolyl)-1-(P-Chlorophenyl)Methane, Modulates Glial Reactivity and Is Neuroprotective in MPTP-Indu
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Molecular, Genetic, and Nutritional Aspects of Major and Trace Minerals
Molecular, Genetic, and Nutritional Aspects of Major and Trace Minerals Edited by James F. Collins AMSTERDAM • BOSTON • HEIDELBERG • LONDON • NEW ORK • OFORD • PARIS SAN DIEGO • SAN FRANCISCO • SINGAPORE • SDNE • TOKO Academic Press is an imprint of Elsevier Academic Press is an imprint of Elsevier 125 London Wall, London EC2Y 5AS, United Kingdom 525 B Street, Suite 1800, San Diego, CA 92101-4495, United States 50 Hampshire Street, 5th Floor, Cambridge, MA 02139, United States The Boulevard, Langford Lane, Kidlington, Oxford OX5 1GB, United Kingdom Copyright © 2017 Elsevier Inc. All rights reserved. No part of this publication may be reproduced or transmitted in any form or by any means, electronic or mechanical, including photocopying, recording, or any information storage and retrieval system, without permission in writing from the publisher. Details on how to seek permission, further information about the Publisher’s permissions policies and our arrangements with organizations such as the Copyright Clearance Center and the Copyright Licensing Agency, can be found at our website: www.elsevier.com/permissions. This book and the individual contributions contained in it are protected under copyright by the Publisher (other than as may be noted herein). Notices Knowledge and best practice in this field are constantly changing. As new research and experience broaden our understanding, changes in research methods, professional practices, or medical treatment may become necessary. Practitioners and researchers must always rely on their own experience and knowledge in evaluating and using any information, methods, compounds, or experiments described herein. In using such information or methods they should be mindful of their own safety and the safety of others, including parties for whom they have a professional responsibility. -

Assessment of NR4A Ligands That Directly Bind and Modulate the Orphan Nuclear Receptor Nurr1
bioRxiv preprint doi: https://doi.org/10.1101/2020.05.22.109017; this version posted May 25, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Assessment of NR4A Ligands that Directly Bind and Modulate the Orphan Nuclear Receptor Nurr1 Paola Munoz-Tello 1, Hua Lin 2,3, Pasha Khan 2, Ian Mitchelle S. de Vera 1,4, Theodore M. Kamenecka 2, and Douglas J. Kojetin 1,2,* 1 Department of Integrative Structural and Computational Biology, The Scripps Research Institute, Jupiter, FL, 33458, USA 2 Department of Molecular Medicine, The Scripps Research Institute, Jupiter, Florida 33458, USA 3 Current address: Biomedical Research Center of South China, College of Life Sciences, Fujian Normal University, Fuzhou 350117, China 4 Current address: Department of Pharmacology and Physiology, Saint Louis University School of Medicine, St. Louis, MO 63104, USA * Correspondence: [email protected] as ligands that increase Nurr1 transcription in human neuroblastoma SK- ABSTRACT N-BE(2)-C cells (11). Amodiaquine improves behavioral alterations in a Nurr1/NR4A2 is an orphan nuclear receptor transcription factor im- Parkinson’s disease animal model (11) and improves neuropathology and plicated as a potential drug target for neurological disorders including memory impairment in an Alzheimer’s disease animal model (12). Alzheimer’s and Parkinson’s diseases. Previous studies identified small molecule modulators of NR4A nuclear receptors including Nurr1 and Amodiaquine is the most potent and efficacious Nurr1 agonist of the Nur77/NR4A1; it remains unclear whether these ligands affect Nurr1 4-amino-7-chloroquinoline compounds, but these compounds are not through direct binding or indirect non-binding mechanisms. -

Table 2. Significant
Table 2. Significant (Q < 0.05 and |d | > 0.5) transcripts from the meta-analysis Gene Chr Mb Gene Name Affy ProbeSet cDNA_IDs d HAP/LAP d HAP/LAP d d IS Average d Ztest P values Q-value Symbol ID (study #5) 1 2 STS B2m 2 122 beta-2 microglobulin 1452428_a_at AI848245 1.75334941 4 3.2 4 3.2316485 1.07398E-09 5.69E-08 Man2b1 8 84.4 mannosidase 2, alpha B1 1416340_a_at H4049B01 3.75722111 3.87309653 2.1 1.6 2.84852656 5.32443E-07 1.58E-05 1110032A03Rik 9 50.9 RIKEN cDNA 1110032A03 gene 1417211_a_at H4035E05 4 1.66015788 4 1.7 2.82772795 2.94266E-05 0.000527 NA 9 48.5 --- 1456111_at 3.43701477 1.85785922 4 2 2.8237185 9.97969E-08 3.48E-06 Scn4b 9 45.3 Sodium channel, type IV, beta 1434008_at AI844796 3.79536664 1.63774235 3.3 2.3 2.75319499 1.48057E-08 6.21E-07 polypeptide Gadd45gip1 8 84.1 RIKEN cDNA 2310040G17 gene 1417619_at 4 3.38875643 1.4 2 2.69163229 8.84279E-06 0.0001904 BC056474 15 12.1 Mus musculus cDNA clone 1424117_at H3030A06 3.95752801 2.42838452 1.9 2.2 2.62132809 1.3344E-08 5.66E-07 MGC:67360 IMAGE:6823629, complete cds NA 4 153 guanine nucleotide binding protein, 1454696_at -3.46081884 -4 -1.3 -1.6 -2.6026947 8.58458E-05 0.0012617 beta 1 Gnb1 4 153 guanine nucleotide binding protein, 1417432_a_at H3094D02 -3.13334396 -4 -1.6 -1.7 -2.5946297 1.04542E-05 0.0002202 beta 1 Gadd45gip1 8 84.1 RAD23a homolog (S. -

Supplementary Data Genbank Or OSE Vs RO NIA Accession Gene Name Symbol FC B-Value H3073C09 11.38 5.62 H3126B09 9.64 6.44 H3073B0
Supplementary Data GenBank or OSE vs RO NIA accession Gene name Symbol FC B-value H3073C09 11.38 5.62 H3126B09 9.64 6.44 H3073B08 9.62 5.59 AU022767 Exportin 4 Xpo4 9.62 6.64 H3073B09 9.59 6.48 BG063925 Metallothionein 2 Mt2 9.23 18.89 H3064B07 9.21 6.10 H3073D08 8.28 6.10 AU021923 Jagged 1 Jag1 7.89 5.93 H3070D08 7.54 4.58 BG085110 Cysteine-rich protein 1 (intestinal) Crip1 6.23 16.40 BG063004 Lectin, galactose binding, soluble 1 Lgals1 5.95 10.36 BG069712 5.92 2.34 BG076976 Transcribed locus, strongly similar to NP_032521.1 lectin, galactose binding, soluble 1 5.64 8.36 BG062930 DNA segment, Chr 11, Wayne State University 99, expressed D11Wsu99e 5.63 8.76 BG086474 Insulin-like growth factor binding protein 5 Igfbp5 5.50 15.95 H3002d11 5.13 20.77 BG064706 Keratin complex 1, acidic, gene 19 Krt1-19 5.06 9.07 H3007A09 5.05 2.46 H3065F02 4.84 5.43 BG081752 4.81 1.25 H3010E09 4.71 11.90 H3064c11 4.43 1.00 BG069711 Transmembrane 4 superfamily member 9 Tm4sf9 4.29 1.23 BG077072 Actin, beta, cytoplasmic Actb 4.29 3.01 BG079788 Hemoglobin alpha, adult chain 1 Hba-a1 4.26 6.63 BG076798 4.23 0.80 BG074344 Mesothelin Msln 4.22 6.97 C78835 Actin, beta, cytoplasmic Actb 4.16 3.02 BG067531 4.15 1.61 BG073468 Hemoglobin alpha, adult chain 1 Hba-a1 4.10 6.23 H3154H07 4.08 5.38 AW550167 3.95 5.66 H3121B01 3.94 5.94 H3124f12 3.94 5.64 BG073608 Hemoglobin alpha, adult chain 1 Hba-a1 3.84 5.32 BG073617 Hemoglobin alpha, adult chain 1 Hba-a1 3.84 5.75 BG072574 Hemoglobin alpha, adult chain 1 Hba-a1 3.82 5.93 BG072211 Tumor necrosis factor receptor superfamily, -

Rapid Effects of LH on Gene Expression in the Mural Granulosa Cells of Mouse Periovulatory Follicles
REPRODUCTIONRESEARCH Rapid effects of LH on gene expression in the mural granulosa cells of mouse periovulatory follicles Martha Z Carletti and Lane K Christenson Department of Molecular and Integrative Physiology, University of Kansas Medical Center, 3075 KLSIC, 3901 Rainbow Boulevard, Kansas City, Kansas 66160, USA Correspondence should be addressed to L K Christenson; Email: [email protected] Abstract LH acts on periovulatory granulosa cells by activating the PKA pathway as well as other cell signaling cascades to increase the transcription of specific genes necessary for ovulation and luteinization. Collectively, these cell signaling responses occur rapidly (within minutes); however, presently no high throughput studies have reported changes before 4 h after the LH surge. To identify early response genes that are likely critical for initiation of ovulation and luteinization, mouse granulosa cells were collected before and 1 h after hCG. Fifty-seven gene transcripts were significantly (P!0.05) upregulated and three downregulated following hCG. Twenty-four of these transcripts were known to be expressed after the LH/hCG surge at later time points, while 36 were unknown to be expressed by periovulatory granulosa cells. Temporal expression of several transcripts, including the transcription factors Nr4a1, Nr4a2, Egr1, Egr2, Btg1, and Btg2, and the epidermal growth factor (EGF)-like ligands Areg and Ereg, were analyzed by quantitative RT-PCR, and their putative roles in granulosa cell function are discussed. Epigen (Epgn), another member of the family of EGF-like ligands was identified for the first time in granulosa cells as rapidly induced by LH/hCG. We demonstrate that Epgn initiates cumulus expansion, similar to the other EGF-receptor ligands Areg and Ereg. -

Imbim Annual Report 2015
Department of Medical Biochemistry and Microbiology IMBIM ANNUAL REPORT 2015 DEPARTMENT OF MEDICAL BIOCHEMISTRY AND MICROBIOLOGY ANNUAL REPORT 2015 Pictures taken by Helena Öhrvik The role of copper in mast cell granule homeostasis Ctr2 is a protein involved in cellular transport of copper. Upper panels: staining for tryptase, a mast cell granule protease, in tryptase (Mcpt6)-deficient (negative control), Ctr2-/- and wild type (WT) mast cells. Note that the absence of Ctr2 causes upregulated expression of tryptase. Lower panels: staining of Ctr2-/- and WT mast cells with toluidine blue, a proteoglycan- binding dye. Note increased toluidine blue staining of Ctr2-/- mast cells, indicating increased proteoglycan content of granules. Öhrvik, H., Logeman, B., Noguchi, G., Eriksson, I., Kjellén, L., Thiele, D.J., Pejler, G. (2015) Ctr2 regulates mast cell maturation by affecting the storage and expression of tryptase and proteoglycans. J. Immunol. 195, 3654-364. Edited by Veronica Hammar ISBN no 978-91-979531-8-4 PREFACE Another year has passed with both small and large successes for the laboratory. The Department has a stable staff consisting of some 150 people. Including project workers, post doc etc we are around 250 persons that spend our daily work hours at the Department. Fortunately the granting situation for the IMBIM researchers improved considerably during 2015. Many of the small and medium sized research groups received grants that will help them continue to excel during the coming years. In addition Dan Andersson, Leif Andersson, Per Jemth, and a couple of scientists at ICM, received a hefty 47 milj kr grant from “Knut och Alice Wallenbergs Stiftelse” to support their research on the evolution of new genes and proteins. -

Of the T Cell by G-CSF and IL-10 Cell Mobilization Requires Direct Signaling Modification of T Cell Responses by Stem
Downloaded from http://www.jimmunol.org/ by guest on September 29, 2021 is online at: average * The Journal of Immunology published online 28 February 2014 from submission to initial decision 4 weeks from acceptance to publication http://www.jimmunol.org/content/early/2014/02/28/jimmun ol.1302315 Modification of T Cell Responses by Stem Cell Mobilization Requires Direct Signaling of the T Cell by G-CSF and IL-10 Kelli P. A. MacDonald, Laetitia Le Texier, Ping Zhang, Helen Morris, Rachel D. Kuns, Katie E. Lineburg, Lucie Leveque, Alistair L. Don, Kate A. Markey, Slavica Vuckovic, Frederik O. Bagger, Glen M. Boyle, Bruce R. Blazar and Geoffrey R. Hill J Immunol Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2014/02/28/jimmunol.130231 5.DCSupplemental Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2014 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 29, 2021. Published February 28, 2014, doi:10.4049/jimmunol.1302315 The Journal of Immunology Modification of T Cell Responses by Stem Cell Mobilization Requires Direct Signaling of the T Cell by G-CSF and IL-10 Kelli P. -

3,3'-Diindolylmethane and Its Derivatives: Nature
Biersack. Cancer Drug Resist 2020;3:867-78 Cancer DOI: 10.20517/cdr.2020.53 Drug Resistance Review Open Access 3,3’-Diindolylmethane and its derivatives: nature- inspired strategies tackling drug resistant tumors by regulation of signal transduction, transcription factors and microRNAs Bernhard Biersack Organic Chemistry 1, University of Bayreuth, Bayreuth 95440, Germany. Correspondence to: Dr. Bernhard Biersack, Organic Chemistry 1, University of Bayreuth, Universitätsstrasse 30, Bayreuth 95440, Germany. E-mail: [email protected] How to cite this article: Biersack B. 3,3’-Diindolylmethane and its derivatives: nature-inspired strategies tackling drug resistant tumors by regulation of signal transduction, transcription factors and microRNAs. Cancer Drug Resist 2020;3:867-78. http://dx.doi.org/10.20517/cdr.2020.53 Received: 21 Jul 2020 First Decision: 24 Aug 2020 Revised: 31 Aug 2020 Accepted: 22 Sep 2020 Available online: 12 Oct 2020 Academic Editor: Shrikant Anant Copy Editor: Cai-Hong Wang Production Editor: Jing Yu Abstract Indoles of cruciferous vegetables are promising anti-tumor agents. Studies with indole-3-carbinol and its dimeric product, 3,3’-diindolylmethane (DIM), suggest that these compounds have the ability to deregulate multiple cellular signaling pathways that are essential for tumor growth and spread. These natural compounds are also effective modulators of transcription factors and non-coding RNAs. These effects explain their ability to inhibit tumor spread and to overcome drug resistance. In this work, pertinent literature on the effects of DIM and its synthetic derivatives on resistant tumors and resistance mechanisms in tumors is highlighted. Keywords: Indole, 3,3’-diindolylmethane, drug resistance, anticancer drugs, microRNAs INTRODUCTION Cancer is one of the most serious human health concerns. -

The Molecular Mechanisms by Which Vitamin D Prevents Insulin Resistance and Associated Disorders
International Journal of Molecular Sciences Review The Molecular Mechanisms by Which Vitamin D Prevents Insulin Resistance and Associated Disorders Izabela Szymczak-Pajor 1 ,Józef Drzewoski 2 and Agnieszka Sliwi´ ´nska 1,* 1 Department of Nucleic Acid Biochemistry, Medical University of Lodz, 251 Pomorska Str., 92-213 Lodz, Poland; [email protected] 2 Central Teaching Hospital of the Medical University of Lodz, 251 Pomorska Str., 92-213 Lodz, Poland; [email protected] * Correspondence: [email protected] Received: 9 August 2020; Accepted: 9 September 2020; Published: 11 September 2020 Abstract: Numerous studies have shown that vitamin D deficiency is very common in modern societies and is perceived as an important risk factor in the development of insulin resistance and related diseases such as obesity and type 2 diabetes (T2DM). While it is generally accepted that vitamin D is a regulator of bone homeostasis, its ability to counteract insulin resistance is subject to debate. The goal of this communication is to review the molecular mechanism by which vitamin D reduces insulin resistance and related complications. The university library, PUBMED, and Google Scholar were searched to find relevant studies to be summarized in this review article. Insulin resistance is accompanied by chronic hyperglycaemia and inflammation. Recent studies have shown that vitamin D exhibits indirect antioxidative properties and participates in the maintenance of normal resting ROS level. Appealingly, vitamin D reduces inflammation and regulates Ca2+ level in many cell types. Therefore, the beneficial actions of vitamin D include diminished insulin resistance which is observed as an improvement of glucose and lipid metabolism in insulin-sensitive tissues. -

Supplemental Materials
The infection-tolerant mammalian reservoir of Lyme disease and other zoonoses broadly counters the inflammatory effects of endotoxin Supplemental Materials Figures S1-S5 Tables S1-S20 Figure S1. Digital photograph of two adult Peromyscus leucopus with exudative conjunctivitis and huddled together. The animals had received 10 mg/gm body of Escherichia coli lipopolysaccharide intraperitoneally the day before. Figure S2. Species- and tissue-specific responses to LPS. Independent differential gene expression analysis of RNA-seq data were performed for blood, spleen, and liver tissues of P. leucopus and M. musculus collected 4 h after injection with LPS or buffer alsone as control. These are represented as volcano plots with range- adjusted scales for the log2-transformed fold-changes on x-axes and log10-transformed FDR p values on y- axes. Colors of symbols denote the following: red, up-regulated gene with absolute fold-change > 4.0 and p value < 0.05; purple, down-regulated gene with absolute fold-change > 4.0 and p value < 0.05; and gray, all others. Numbers at the top left and right corners in each plot respresent numbers of down- and up-regulated genes, respectively. Figure 3 is same data with constant scales for x- and y-axes across the plots. Numerical values for each gene in the 6 datasets are provided in Tables S4-S9. Figure S3. Correlation of IL-10 and IL-10 P. leucopus and M. musculus from RNA-seq of spleen and of Figure 6B and TaBle S14. The scatter plot is log10 values of normalized unique reads of one coding sequence against another for each of the four groups, as defined in the legend for Figure 6 and indicated By different symBols. -
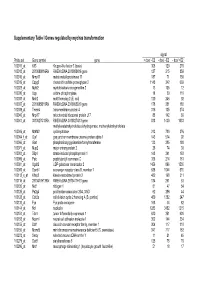
Supplementary Table I Genes Regulated by Myc/Ras Transformation
Supplementary Table I Genes regulated by myc/ras transformation signal Probe set Gene symbol gene + dox/ - E2 - dox/ - E2 - dox/ +E2 100011_at Klf3 Kruppel-like factor 3 (basic) 308 120 270 100013_at 2010008K16Rik RIKEN cDNA 2010008K16 gene 127 215 859 100016_at Mmp11 matrix metalloproteinase 11 187 71 150 100019_at Cspg2 chondroitin sulfate proteoglycan 2 1148 342 669 100023_at Mybl2 myeloblastosis oncogene-like 2 13 105 12 100030_at Upp uridine phosphorylase 18 39 110 100033_at Msh2 mutS homolog 2 (E. coli) 120 246 92 100037_at 2310005B10Rik RIKEN cDNA 2310005B10 gene 178 351 188 100039_at Tmem4 transmembrane protein 4 316 135 274 100040_at Mrpl17 mitochondrial ribosomal protein L17 88 142 89 100041_at 3010027G13Rik RIKEN cDNA 3010027G13 gene 818 1430 1033 methylenetetrahydrofolate dehydrogenase, methenyltetrahydrofolate 100046_at Mthfd2 cyclohydrolase 213 720 376 100064_f_at Gja1 gap junction membrane channel protein alpha 1 143 574 92 100066_at Gart phosphoribosylglycinamide formyltransferase 133 385 180 100071_at Mup2 major urinary protein 2 26 74 30 100081_at Stip1 stress-induced phosphoprotein 1 148 341 163 100089_at Ppic peptidylprolyl isomerase C 358 214 181 100091_at Ugalt2 UDP-galactose translocator 2 1456 896 1530 100095_at Scarb1 scavenger receptor class B, member 1 638 1044 570 100113_s_at Kifap3 kinesin-associated protein 3 482 168 311 100116_at 2810417H13Rik RIKEN cDNA 2810417H13 gene 134 261 53 100120_at Nid1 nidogen 1 81 47 54 100125_at Pa2g4 proliferation-associated 2G4, 38kD 62 286 44 100128_at Cdc2a cell division cycle 2 homolog A (S. pombe) 459 1382 247 100133_at Fyn Fyn proto-oncogene 168 44 82 100144_at Ncl nucleolin 1283 3452 1215 100151_at Tde1 tumor differentially expressed 1 620 351 620 100153_at Ncam1 neural cell adhesion molecule 1 302 144 234 100155_at Ddr1 discoidin domain receptor family, member 1 304 117 310 100156_at Mcmd5 mini chromosome maintenance deficient 5 (S. -

Rabbit Anti-EWSR1 Polyclonal Antibody Reactivity: Hu, Rt, Ms, Dg Synonym: Ewing Sarcoma Breakpoint Region 1, EWS, EWSR1, EWSR-1
Order Information Description: Rabbit anti-EWSR1 Catalogue#: 600-820 Lot#: See the label Size: 100 ug/200 ul Host: Rabbit Clone: N/A Application: ELISA, WB, IHC, Rabbit anti-EWSR1 Polyclonal Antibody Reactivity: Hu, Rt, Ms, Dg Synonym: Ewing sarcoma breakpoint region 1, EWS, EWSR1, EWSR-1. ANTIGEN PREPARATION A synthetic peptide corresponding to the internal segment of EWSR1 protein. This sequence is identical among human, rat, mouse and dog species. BACKGROUND The EWSR1 gene encodes a putative RNA binding protein. Mutation in this gene causes Ewing sarcoma and neuroectodermal and various other tumors. This gene is involved in a number of different sarcomas as a result of chromosomal translocations. EWSR1 is fused to an ETS transcription factor (FLI-1, ERG, ETV-1, E1AF or FEV) in Ewing sarcoma; to activating transcription factor-1 (ATF-1) in soft tissue clear cell sarcoma; to Wilms Tumor -1 (WT1) in desmoplastic small round cell; to nuclear receptor 4A3 (NR4A3) in extraskeletal myxoid chondrosarcoma, and to C/EBP- homologous protein (CHOP) in myxoid liposarcoma. EWSR1 genes are required for mitotic integrity and survival of neural cells in the early embryonic development, and that the involvement of EWSR1 in mitotic stability is conserved from zebrafish to human. PURIFICATION FORMULATION This affinity purified antibody is supplied in sterile phosphate- The Rabbit IgG is purified by Epitope Affinity Purification. buffered saline (pH7.2) containing antibody stabilizer SPECIFICITY STORAGE This antibody recognizes ~70 kDa of EWSR1 protein. It The antibodies are stable for 12 months from date of receipt when also cross reacts with human, mouse and rat tissues.