Research of 5 Extracts of Wild Amur Grape Vitis Amurensis Rupr. and Identification of Its Polyphenolic Composition by Tandem Mass Spectrometry (HPLC-MS / MS)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
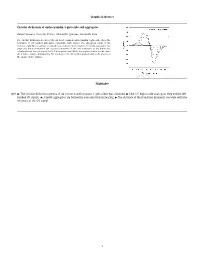
Graphical Abstract Circular Dichroism of Anthocyanidin 3-Glucoside Self
Graphical abstract Circular dichroism of anthocyanidin 3-glucoside self-aggregates Raquel Gavara, Vesselin Petrov, Alexandre Quintas, Fernando Pina * The circular dichroism spectra of the six most common anthocyanidin 3-glucoside show the formation of left handed aggregates compatible with dimers. The absorption bands of the monomer split by increasing concentration according to the formation of H and J aggregates. The angle and distance between the transition moments of the two monomers in the dimer was calculated from the splitting of the 0–0 absorption band. While the angle is similar for the series the distance changes dramatically. The intensity of the CD signal is proportional to the inverse of the square of the distance. Highlights Q10 " The circular dichroism spectra of six common anthocyanins 3-glucosides was obtained. " Like 3,5-diglucoside analogous they exhibit left- handed CD signals. " J and H aggregates are formed by concentration increasing. " The distance of the transition moments correlate with the intensity of the CD signal. 1 1 2 Circular dichroism of anthocyanidin 3-glucoside self-aggregates a a b a,⇑ 3 Q1 Raquel Gavara , Vesselin Petrov , Alexandre Quintas , Fernando Pina 4 a REQUIMTE, Departamento de Química, Faculdade de Ciências e Tecnologia, Universidade Nova de Lisboa, 2829516 Monte de Caparica, Portugal 5 b Instituto Superior de Ciencias da Saude Egas Moniz, Centro de Investigação Interdisciplinar Egas Moniz, P-2829511 Monte de Caparica, Caparica, Portugal 6 7 article info a b s t r a c t 1 9 10 Self-association constants for the flavylium cations of the six most common anthocyanidin 3-glucosides 20 11 were determined by circular dichroism (CD) and UV–Vis spectroscopy. -

Electrochemical and Antioxidant Properties of Anthocyanins and Anthocyanidins
CROATICA CHEMICA ACTA CCACAA 80 (1) 29¿34 (2007) ISSN-0011-1643 CCA-3135 Original Scientific Paper Electrochemical and Antioxidant Properties of Anthocyanins and Anthocyanidins Andréia A. de Lima, Eliana M. Sussuchi, and Wagner F. De Giovani* Departamento de Química, Faculdade de Filosofia, Ciências e Letras de Ribeirão Preto, Universidade de São Paulo, Av. Bandeirantes 3900, 14040-901, Ribeirão Preto – SP, Brazil RECEIVED JUNE 26, 2006; REVISED SEPTEMBER 29, 2006; ACCEPTED OCTOBER 27, 2006 Electrochemical properties of delphinidin, cyanidin, pelargonidin, kuromanin and callistephin were investigated by cyclic and differential pulse voltammetries at different pH values and also in methanol. On the basis of oxidation potentials, the order of antioxidant activity for antho- cyanidins is delphinidin > cyanidin > pelargonidin. Oxidation peaks for anthocyanins (kuro- manin and callistephin) are shifted to more positive potentials compared to anthocyanidins (delphinidin, cyanidin and pelargonidin). Oxidation peak currents are linearly dependent on the Keywords square root of the scan rate, which is typical of a diffusion controlled electrochemical process. anthocyanins Antioxidant activities of the compounds were evaluated using the 1,1-diphenyl-2-picrylhydra- anthocyanidins zyl (DPPH) radical-scavenging method and they are directly related to their redox potential antioxidants values. The order of the antioxidant activity is delphinidin > cyanidin > pelargonidin > kuro- electrochemical properties manin > callistephin. INTRODUCTION pharmacological properties that render them interesting potential cancer chemopreventive agents.9–11 Because Anthocyanins are one of the main classes of flavonoids anthocyanins are widely consumed, finding out addi- in red wines. They have been pointed out as the molecu- tional biological activities related to these compounds lar entities that are most likely responsible for what has would be of great interest. -

Callistephin Enhances the Protective Effects of Isoflurane on Microglial Injury Through Downregulation of Inflammation and Apoptosis
802 MOLECULAR MEDICINE REPORTS 20: 802-812, 2019 Callistephin enhances the protective effects of isoflurane on microglial injury through downregulation of inflammation and apoptosis LILI ZHAO, SHIBIAO CHEN, TIANYIN LIU, XIUHONG WANG, HAIJIN HUANG and WEICHENG LIU Department of Anesthesiology, The First Affiliated Hospital of Nanchang University, Nanchang, Jiangxi 330006, P.R. China Received June 18, 2018; Accepted March 15, 2019 DOI: 10.3892/mmr.2019.10282 Abstract. Microglia are the major immune cells in the central enhanced the effects of isoflurane. Callistephin may therefore nervous system. Microglial activation can be beneficial or constitute a candidate drug agent that may target inflammatory detrimental depending on the stimuli and the physiopathological and growth regulatory signaling pathways, thus ameliorating environment. Microglial activation is involved in a variety certain aspects of neurodegenerative diseases. of neurodegenerative disorders. Different anesthetic agents have exhibited diverse effects on microglial activation and Introduction the engulfment process. The anthocyanin callistephin has been demonstrated to have antioxidant and anti‑inflammatory Microglial cells are the major immune cell in the central properties, and these were assessed in the present study, with a nervous system (CNS), responding against types of endog- focus on its effect on microglial activation. Mouse microglial enous and exogenous stimuli, including infection by bacteria, cells C8-4B were treated with 100 ng/µl lipopolysaccharide viruses, prions and β-amyloid plaques (1). Microglia are (LPS) and 1 ng/µl interferon-γ. Cells were subsequently treated activated upon exposure to different stimuli and, depending with 2% isoflurane, 100 µM callistephin or both. LPS promoted on the environmental context, this may be beneficial or detri- apoptosis in C8-B4 cells, and this was reduced following mental to the functionality and physiology of the CNS (2). -

Characterisation of Bioactive Compounds in Berries from Plants Grown Under Innovative Photovoltaic Greenhouses
Journal of Berry Research 8 (2018) 55–69 55 DOI:10.3233/JBR-170258 IOS Press Characterisation of bioactive compounds in berries from plants grown under innovative photovoltaic greenhouses Federica Blandoa,∗, Carmela Gerardia, Massimiliano Rennab,c, Sergio Castellanod and Francesco Seriob aInstitute of Sciences of Food Production (ISPA), CNR, Lecce, Italy bInstitute of Sciences of Food Production (ISPA), CNR, Bari, Italy cDepartment of Agricultural and Environmental Science, University of Bari Aldo Moro, Bari, Italy dDepartment of Science of Agriculture, Food and Environment, (SAFE) University of Foggia, Foggia, Italy Received 2 July 2017; accepted 1 November 2017 Abstract. BACKGROUND: Bioactive compounds, mainly polyphenols, present in berries, are thought to be responsible for the health benefits of these fruit. Therefore, it is worthwhile to define the optimal environmental conditions to maximise their polyphenol content. OBJECTIVE: With the aim to define the optimal conditions for berry cultivation in an innovative environment, red rasp- berry, wild strawberry and blackberry plants were grown in a traditional greenhouse in comparison with two photovoltaic greenhouses with different shading area. METHODS: Hydroalcoholic extracts of ripe berries were evaluated by HPLC analysis, for their anthocyanins, organic acids and sugar contents. Moreover, phenolic content (by the Folin-Ciocalteu assay) and antioxidant activity (by the Trolox equivalent antioxidant capacity-TEAC assay) were assayed on the same berry extracts. RESULTS: Total anthocyanins, -

First-Pass Metabolism of Polyphenols from Selected Berries: a High-Throughput Bioanalytical Approach
antioxidants Article First-Pass Metabolism of Polyphenols from Selected Berries: A High-Throughput Bioanalytical Approach Francisco J. Olivas-Aguirre 1,*, Sandra Mendoza 2, Emilio Alvarez-Parrilla 3 , Gustavo A. Gonzalez-Aguilar 4 , Monica A. Villegas-Ochoa 4 , Jael T.J. Quintero-Vargas 1 and Abraham Wall-Medrano 3,* 1 Departamento de Ciencias de la Salud, Universidad de Sonora (Campus Cajeme), Blvd Bordo Nuevo s/n, Ejido Providencia, Cd, Obregón 85199, Mexico; [email protected] 2 Departamento de Investigación y Posgrado en Alimentos (PROPAC), Facultad de Química, Universidad Autónoma de Querétaro, Cerro de las Campanas s/n, Santiago de Querétaro 76010, Mexico; [email protected] 3 Instituto de Ciencias Biomédicas, Universidad Autónoma de Ciudad Juárez, Anillo Envolvente del PRONAF y Estocolmo s/n, Ciudad Juárez 32310, Mexico; [email protected] 4 Centro de Investigación en Alimentación y Desarrollo, A.C. (CIAD), Carretera a Ejido La Victoria, Km. 0.6, Hermosillo 83304, Mexico; [email protected] (G.A.G.-A.); [email protected] (M.A.V.-O.) * Correspondence: [email protected] (F.J.O.-A.); [email protected] (A.W.-M.) Received: 29 March 2020; Accepted: 11 April 2020; Published: 13 April 2020 Abstract: Small berries are rich in polyphenols whose first-pass metabolism may alter their ultimate physiological effects. The antioxidant capacity and polyphenol profile of three freeze-dried berries (blackberry, raspberry, Red Globe grape) were measured and their apparent permeability (Papp) and first-pass biotransformation were tracked with an ex vivo bioanalytical system [everted gut sac (rat) + three detection methods: spectrophotometry, HPLC-ESI-QTOF-MS, differential pulse voltammetry (DPV)]. -

Effect of Different Anthocyanidin Glucosides on Lutein Uptake By
molecules Article Effect of Different Anthocyanidin Glucosides on Lutein Uptake by Caco-2 Cells, and Their Combined Activities on Anti-Oxidation and Anti-Inflammation In Vitro and Ex Vivo Minh Anh Thu Phan 1,*, Martin Bucknall 2 and Jayashree Arcot 1,* ID 1 Food and Health Cluster, School of Chemical Engineering, UNSW, Sydney, NSW 2052, Australia 2 Mark Wainwright Analytical Centre, UNSW, Sydney, NSW 2052, Australia; [email protected] * Correspondence: [email protected] (M.A.T.P.); [email protected] (J.A.); Tel.: +61-2-9385-5360 (J.A.) Academic Editors: Natália Martins and Derek J. McPhee Received: 23 July 2018; Accepted: 11 August 2018; Published: 14 August 2018 Abstract: The interactive effects on anti-oxidation and anti-inflammation of lutein combined with each of the six common anthocyanidin glucosides were studied in both chemical and cellular systems. The combined phytochemicals showed an antagonism in the inhibition of lipid oxidation in a liposomal membrane, but showed an additive effect on cellular antioxidant activity in Caco-2 cells. Lutein was an active lipoxygenase inhibitor at 2–12 µM while anthocyanins were inactive. The concentration of lutein when it was used in combination with anthocyanins was 25–54% higher than when lutein was used alone (i.e., IC50 = 1.2 µM) to induce 50% of lipoxygenase inhibition. Only the combination of lutein with malvidin-3-glucoside showed anti-inflammatory synergy in the suppression of interleukin-8, and the synergy was seen at all three ratios tested. Some mixtures, however, showed anti-inflammatory antagonism. The presence of anthocyanins (5–7.5 µM) did not affect lutein uptake (2.5–5 µM) by Caco-2 cells. -

Changes in Phenolic Compounds and Antioxidant Activity of Fruit Musts and Fruit Wines During Simulated Digestion
molecules Article Changes in Phenolic Compounds and Antioxidant Activity of Fruit Musts and Fruit Wines during Simulated Digestion Tomasz Tarko * , Aleksandra Duda-Chodak and Agata Soszka Department of Fermentation Technology and Microbiology, Faculty of Food Technology, University of Agriculture in Krakow, ul. Balicka 122, 30-149 Krakow, Poland; [email protected] (A.D.-C.); [email protected] (A.S.) * Correspondence: [email protected]; Tel.: +48-12-6624792 Academic Editor: Urszula Gawlik-Dziki Received: 5 November 2020; Accepted: 24 November 2020; Published: 27 November 2020 Abstract: The content of polyphenols (total phenolic content (TPC)) and the antioxidant activity (AOX) of food products depend on the raw materials used and the technological processes in operation, but transformations of these compounds in the digestive tract are very important. The aim of this study was to determine the TPC, profile of polyphenols, and AOX of apple and blackcurrant musts and wines in order to evaluate the changes occurring in a simulated human digestive system. The research material consisted of apples and blackcurrant, from which musts and fruit wines were obtained. All samples were subjected to three-stage digestion in a simulated human digestive system and then analyzed for the following: TPC (Folin–Ciocalteu method) and profile (HPLC), AOX (method with 2,20-azino-bis (3-ethylbenzothiazoline-6-sulfonic) acid (ABTS) radical), and for the wines also total extract, volatile acidity (International Organization of Vine and Wine (OIV) method), and sugar profile (HPLC). The antioxidant activity of fruit wines is directly related to the total polyphenol content. Phenolic compounds were transformed during all digestive stages, which led to the formation of compounds with higher antioxidant capacity. -

Note Formation of an Inclusion Complex of Anthocyanin With
Agric. Biol. Chem., 44 (6), 1411•`1413 , 1980 1411 Note extracted from the seeds of black soy bean (KUROMAME) (Glycine max Merill). Nasunin was ex tracted from the fruits of egg plant (Solanum melongena L .). Formation of an Inclusion Complex These three anthocyanins extracted were proved to be pure of Anthocyanin with Cyclodextrin by paper chromatography after some preparation procedure.6) ƒ¿-CD and ƒÀ-CD used were commercially available Tetsuya YAMADA,Takashi KOMIYA (Hayashibara Biochemical Research), and amylose was and Morio AKAKI prepared from a high amylose corn starch. Amylose was dissolved in dimethylsulfoxide (DMSO) and the solution Facultyof Agriculture,Mie University, was diluted with water, final concentration of DMSO being Tsu 514,Japan 33%. ReceivedJuly 24, 1979 Each anthocyanin was dissolved in water, further diluted properly with water and adjusted to pH 2.0 with HCl. To each of these anthocyanin solutions was added cyclodextrin solution or amylose solution at various concentrations. Recently, many studies concerning the inclusion complex Addition of ƒÀ-CD resulted in the fading of two an forming activity of cyclodextrin were reported .1•`3) We thocyanin solutions (callistephin and chrysanthemin), and were also interested in this activity because cyclodextrin is this fading effect was greater with higher concentrations of analogous to amylose and amylopectin. Since many natural -CD added (Table I) . On the other hand, addition of ƒ¿-CD ƒÀ pigments in food coexist with saccharides, it was wondered had almost no fading effect in chrysanthemin solution, if the stability of these pigments was affected with the although it showed a less but significant fading effect in presence of such saccharides. -

Color Pigmentation in Dionaea Muscipula Ellis and Drosera Spathulata Labill
235 Short Communication Plant Biotechnology, 16 (3), 235-238 (1999) Effects of Macro-components and Sucrose in the Medium on in vitro Red- color Pigmentation in Dionaea muscipula Ellis and Drosera spathulata Labill. Sayuri ICHIISHI*, Toshiharu NAGAMITSU*, Yusuke KONDO*, Tsukasa IWASHINA* *, Katsuhiko KONDO* * *'t and Norikazu TAGASHIRA* * * *PlantBio-technology Section , Sanmei Electric Co., Ltd., 5-7, Sumiyoshi- Cho, Handa City 475-0862, Japan **Tsukuba Botanical Garden , National Science Museum, 4-1-1, Amakubo, Tsukuba City 305-0005, Japan ***Laboratory of Plant Chromosome and Gene Stock, Faculty of Science, Hiroshima University, 1-4-3 Kagamiyama, Higashi-Hiroshima City 739-8526, Japan E-mail:kko Received 21 September 1998; accepted 15 March 1999 Abstract Effects of the five macro-components and sucrose in half strength MS (1/2 MS) agar medium on red -color pigmentation were studied in the plant bodies of Dionaea muscipula and Drosera spathulata generated from multiple shoots in vitro. In 1/2 MS agar medium modified with 10.31 mM NH4NO3 and 9.40 mM KNO3 and supplemented with 0.75 or 0% sucrose the subcultured plants continuously proliferated by multiple shoots and generated large, green-colored plants, while with dilution of those nitrogen components and increase of sucrose to 1.5% the red-color anthocyanin pigmentation spread from the glands or glandular hairs to the entire leaves and the plant sizes and dry weight decreased in inverse proportion to the depth of red color. The anthocyanin pigments of Dionaea muscipula consisted of delphinidin 3-0-glucoside which was new to the species and cyanidin 3-0-glucoside (chrysan- themin), and those of Drosera spathulata, studied here for the first time, consisted of cyanidin 3,5-di- 0-glucoside (cyanin), cyanidin 3-0-galactoside (idaein), cyanidin 3-0-glucoside, pelargonidin 3-0- galactoside, and pelargonidin 3-0-glucoside (callistephin). -

日汉食品工业词汇/姬德衡,朱蓓薇主编.—2 版.—北京: 中国轻工业出版社,2008.2 Isbn 978-7-5019-6314-0 日汉食品工业词汇 ⅰ
图书在版编目(CIP)数据 日汉食品工业词汇/姬德衡,朱蓓薇主编.—2 版.—北京: 中国轻工业出版社,2008.2 ISBN 978-7-5019-6314-0 日汉食品工业词汇 Ⅰ. 日… Ⅱ.①姬…②朱… Ⅲ.食品工业—词汇—日、汉 Ⅳ.TS2-61 ( 和漢食品工業用語集) 中国版本图书馆 CIP 数据核字(2007)第 202740 号 (第二版) 责任编辑:李亦兵 涂润林 责任终审:唐是雯 封面设计:李秉奇 版式设计:侯春青 出版发行:中国轻工业出版社(北京东长安街 6 号,邮编:100740) 主 编 印 刷:大连海大印刷有限公司 经 销:各地新华书店 版 次:2008 年 2 月第二版第一次印刷 大连工业大学 开 本:787×1092 1/32 印张:38 字 数:1649 千字 印数:2000 姬德衡 朱蓓薇 书 号:ISBN 978-7-5019-6314-0/TS.3677 定价:150 元 读者服务部邮购热线电话:010-65241695 85111729 传真:85111730 发行电话:010-85119845 65128898 传真:85113293 网 址:http://www.chlip.com.cn E-mail:[email protected] 如发现图书残缺请直接与我社读者服务部联系调换 41261K1X201HBW 1 2 图书在版编目(CIP)数据 日汉食品工业词汇/姬德衡,朱蓓薇主编.—2 版.—北京: 中国轻工业出版社,2008.2 ISBN 978-7-5019-6314-0 日汉食品工业词汇 Ⅰ. 日… Ⅱ.①姬…②朱… Ⅲ.食品工业—词汇—日、汉 Ⅳ.TS2-61 ( 和漢食品工業用語集) 中国版本图书馆 CIP 数据核字(2007)第 202740 号 (第二版) 责任编辑:李亦兵 涂润林 责任终审:唐是雯 封面设计:李秉奇 版式设计:侯春青 出版发行:中国轻工业出版社(北京东长安街 6 号,邮编:100740) 主 编 印 刷:大连海大印刷有限公司 经 销:各地新华书店 版 次:2008 年 2 月第二版第一次印刷 大连工业大学 开 本:787×1092 1/32 印张:38 字 数:1649 千字 印数:2000 姬德衡 朱蓓薇 书 号:ISBN 978-7-5019-6314-0/TS.3677 定价:150 元 读者服务部邮购热线电话:010-65241695 85111729 传真:85111730 发行电话:010-85119845 65128898 传真:85113293 网 址:http://www.chlip.com.cn E-mail:[email protected] 如发现图书残缺请直接与我社读者服务部联系调换 41261K1X201HBW 1 2 第 一 版 内容简介 主 编 郭成勋 姬德衡 本词汇是以 1990 年出版的《日汉食品工业 编写人员 词汇》为基础,经修订、补充编写而成。 郭成勋 姬德衡 李玉振 许安邦 全书收词 60000 余条,收词范围涉及粮食、 专业审校人员 油脂、肉乳、禽蛋、果蔬、水产、罐头、焙烤、 (以姓氏笔划为序) 糕点、糖果、冷冻食品、豆制品、面制品、腌渍 王逢寿 尹宗伦 汪克谦 张齐福 徐云鹏 袁思林 戴行钧 品、饮料、制糖、酿酒、制盐、制茶、烟草、调 主 审 味品、方便食品、保健食品、食品添加剂等食品 徐云鹏 行业;收词内容包括食品质量、安全、卫生、营 封面题字〔日〕岡崎嘉平太 责任编辑 熊慧姗 养、保健、食品原料、贮运、保藏、加工、制造、 封面设计 王丽青 机械、设备、分析、检验、食品工程高新技术, 以及与食品有关的物理学、化学、微生物学等方 第 二 -

© 2017 Sage Willow Haggard
© 2017 Sage Willow Haggard COLOR STABILITY AND PHENOLIC COMPOSITION FROM CORN PERICARP EXTRACTS IN A BEVERAGE MODEL AND PROTECTIVE AND ANTI- INFLAMMATORY POTENTIAL OF ANTHOCYANIN-RICH PLANT EXTRACTS ON CELIAC DISEASE IN VITRO BY SAGE WILLOW HAGGARD THESIS Submitted in partial fulfillment of the requirements for the degree of Master of Science in Food Science and Human Nutrition with a concentration in Food Science in the Graduate College of the University of Illinois at Urbana-Champaign, 2017 Urbana, Illinois Master’s Committee: Professor Elvira Gonzalez de Mejia Professor John A. Juvik Professor Kelly Tappenden ABSTRACT Anthocyanins are increasing in popularity as natural colorants but are less stable than synthetic colorants. Celiac disease is an autoimmune disorder that affects the small intestine when gluten, a protein found in wheat, rye and barley is consumed, the immune system responds producing inflammation and inhibiting the absorption of nutrients into the body. The objective of phase I was to evaluate color stability of nine anthocyanin-rich color corn variety pericarp extracts and the objectives of phase II were to evaluate the potential of anthocyanin- rich plant extracts to protect human epithelial cells against a celiac toxic peptide (CTP) and to compare the effect of different anthocyanin extracts on inflammatory proteins using a human inflammation array. In phase I of this study we compared the color stability of nine anthocyanin-rich extracts from different colored corn varieties pericarp that were added to a beverage model and stored at different temperatures for 12 weeks. After 12 weeks at 32 °C, variety (V)6 (High condensed form, high cyanidin-3-O-glucoside (C3G), low pelargonidin (Pg), low peonidin (Pn), low in acylated forms), had the smallest percent decrease in chroma and longest anthocyanin half-life. -

Flower Color Inheritance of Rhynchostylis Gigantea (Lindl.) Ridl
THE AGRICULTURAL SCIENCE SOCIETY OF THAILAND Flower color inheritance of Rhynchostylis gigantea (Lindl.) Ridl. N. Sumaythachotphong1,*, W. Bundithya1,2 and N. Potapohn1 1 Department of Plant and Soil Sciences, Faculty of Agriculture, Chiang Mai University, Chiang Mai 50200, Thailand 2 Center of Excellence on Agricultural Biotechnology, Science and Technology Postgraduate Education and Research Development Office, Commission on Higher Education, Ministry of Education (AG-BIO/PERDO-CHE), Nakhon Pathom 73140, Thailand * Corresponding author: [email protected] Submission: 30 April 2019 Revised: 3 August 2020 Accepted: 11 August 2020 ABSTRACT Rhynchostylis gigantea has been subjected to a conventional breeding program in order to determine genetic inheritance of flower color. Generally, there are three varieties with four different flower patterns, i.e., white, white with red−pink spots, white with red blotches and red. The pure red is called R. gigantea var. rubrum Sagarik whereas the pure white is called R. gigantea var. harrisonianum HolH., and the white with red−pink spots or blotches is called R. gigantea. Recently, another color, orange−peach, has been developed. The objectives of this study were to determine genetic inheritance and conduct chemical component analysis of flower color inR. gigantea. Chemical analysis of all colors was conducted to identify major color components of the flowers using liquid chromatography−mass spectrometry (LC−MS). Three major components, cyanidin, peonidin and delphinidin were found in the red, white with red blotches and white with red−pink spots forms, whereas pelargonidin was found only in the orange−peach flowers. Anthocyanins were not found in the white color flower. Hybridization was carried out in order to determine color inheritance in these four−color forms: white, white with red blotches, white with red−pink spots and red.