Mouse Spata2 Conditional Knockout Project (CRISPR/Cas9)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Small Cell Ovarian Carcinoma: Genomic Stability and Responsiveness to Therapeutics
Gamwell et al. Orphanet Journal of Rare Diseases 2013, 8:33 http://www.ojrd.com/content/8/1/33 RESEARCH Open Access Small cell ovarian carcinoma: genomic stability and responsiveness to therapeutics Lisa F Gamwell1,2, Karen Gambaro3, Maria Merziotis2, Colleen Crane2, Suzanna L Arcand4, Valerie Bourada1,2, Christopher Davis2, Jeremy A Squire6, David G Huntsman7,8, Patricia N Tonin3,4,5 and Barbara C Vanderhyden1,2* Abstract Background: The biology of small cell ovarian carcinoma of the hypercalcemic type (SCCOHT), which is a rare and aggressive form of ovarian cancer, is poorly understood. Tumourigenicity, in vitro growth characteristics, genetic and genomic anomalies, and sensitivity to standard and novel chemotherapeutic treatments were investigated in the unique SCCOHT cell line, BIN-67, to provide further insight in the biology of this rare type of ovarian cancer. Method: The tumourigenic potential of BIN-67 cells was determined and the tumours formed in a xenograft model was compared to human SCCOHT. DNA sequencing, spectral karyotyping and high density SNP array analysis was performed. The sensitivity of the BIN-67 cells to standard chemotherapeutic agents and to vesicular stomatitis virus (VSV) and the JX-594 vaccinia virus was tested. Results: BIN-67 cells were capable of forming spheroids in hanging drop cultures. When xenografted into immunodeficient mice, BIN-67 cells developed into tumours that reflected the hypercalcemia and histology of human SCCOHT, notably intense expression of WT-1 and vimentin, and lack of expression of inhibin. Somatic mutations in TP53 and the most common activating mutations in KRAS and BRAF were not found in BIN-67 cells by DNA sequencing. -
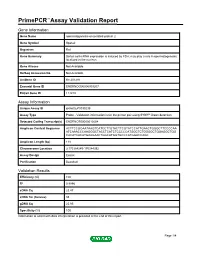
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name spermatogenesis-associated protein 2 Gene Symbol Spata2 Organism Rat Gene Summary Sertoli cell mRNA expression is induced by FSH; may play a role in spermatogenesis; localized to the nucleus Gene Aliases Not Available RefSeq Accession No. Not Available UniGene ID Rn.201291 Ensembl Gene ID ENSRNOG00000009207 Entrez Gene ID 114210 Assay Information Unique Assay ID qRnoCEP0030238 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Coding Transcript(s) ENSRNOT00000012604 Amplicon Context Sequence ACTTCCGGAATAAGTCATCCTTGTACTTCGTATCCATTGAACTGGGCTTCCCCAA ATCAAACCCAAGGGCTACCTCATCTCCCCCATGGCTCTGGGGCTGGAGGCTGG CACATCACATGAAGAACTGGCATGGTGCCCATGGACCAGC Amplicon Length (bp) 118 Chromosome Location 3:170384245-170384392 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 100 R2 0.9996 cDNA Cq 23.47 cDNA Tm (Celsius) 85 gDNA Cq 25.95 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report Spata2, Rat Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. Detected Coding Transcript(s) This is a list of the Ensembl transcript ID(s) that this assay will detect. -

Genetics of Amyotrophic Lateral Sclerosis in the Han Chinese
Genetics of amyotrophic lateral sclerosis in the Han Chinese Ji He A thesis submitted for the degree of Master of Philosophy at The University of Queensland in 2015 The University of Queensland Diamantina Institute 1 Abstract Amyotrophic lateral sclerosis is the most frequently occurring neuromuscular degenerative disorders, and has an obscure aetiology. Whilst major progress has been made, the majority of the genetic variation involved in ALS is, as yet, undefined. In this thesis, multiple genetic studies have been conducted to advance our understanding of the genetic architecture of the disease. In the light of the paucity of comprehensive genetic studies performed in Chinese, the presented study focused on advancing our current understanding in genetics of ALS in the Han Chinese population. To identify genetic variants altering risk of ALS, a genome-wide association study (GWAS) was performed. The study included 1,324 Chinese ALS cases and 3,115 controls. After quality control, a number of analyses were performed in a cleaned dataset of 1,243 cases and 2,854 controls that included: a genome-wide association analysis to identify SNPs associated with ALS; a genomic restricted maximum likelihood (GREML) analysis to estimate the proportion of the phenotypic variance in ALS liability due to common SNPs; and a gene- based analysis to identify genes associated with ALS. There were no genome-wide significant SNPs or genes associated with ALS. However, it was estimated that 17% (SE: 0.05; P=6×10-5) of the phenotypic variance in ALS liability was due to common SNPs. The top associated SNP was within GNAS (rs4812037; p =7×10-7). -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Genome-Wide Pooling Approach Identifies SPATA5 As a New Susceptibility Locus for Alopecia Areata Regina C
Genome-wide pooling approach identifies SPATA5 as a new susceptibility locus for alopecia areata Regina C. Betz, Lina M. Forstbauer, Felix F. Brockschmidt, Valentina Moskvina, Christine Herold, Silke Redler, Alexandra Herzog, Axel M. Hillmer, Christian Meesters, Stefanie Heilmann, et al. To cite this version: Regina C. Betz, Lina M. Forstbauer, Felix F. Brockschmidt, Valentina Moskvina, Christine Herold, et al.. Genome-wide pooling approach identifies SPATA5 as a new susceptibility locus for alopecia areata. European Journal of Human Genetics, Nature Publishing Group, 2011, 10.1038/ejhg.2011.185. hal- 00691329 HAL Id: hal-00691329 https://hal.archives-ouvertes.fr/hal-00691329 Submitted on 26 Apr 2012 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. 1 Genome-wide pooling approach identifies SPATA5 as a new susceptibility locus for 2 alopecia areata 3 4 Lina M. Forstbauer1*, Felix F. Brockschmidt1,2*, Valentina Moskvina3, Christine 5 Herold4, Silke Redler1, Alexandra Herzog1, Axel M. Hillmer5, Christian Meesters4,6, 6 Stefanie Heilmann1,2, Florian Albert1, Margrieta Alblas1,2, Sandra Hanneken7, Sibylle 7 Eigelshoven7, Kathrin A. Giehl8, Dagny Jagielska1,9, Ulrike Blume-Peytavi9, Natalie 8 Garcia Bartels9, Jennifer Kuhn10,11,12, Hans Christian Hennies10,11,12, Matthias 9 Goebeler13, Andreas Jung13, Wiebke K. -

Original Article Identification of Differentially Expressed Genes Between Male and Female Patients with Acute Myocardial Infarction Based on Microarray Data
Int J Clin Exp Med 2019;12(3):2456-2467 www.ijcem.com /ISSN:1940-5901/IJCEM0080626 Original Article Identification of differentially expressed genes between male and female patients with acute myocardial infarction based on microarray data Huaqiang Zhou1,2*, Kaibin Yang2*, Shaowei Gao1, Yuanzhe Zhang2, Xiaoyue Wei2, Zeting Qiu1, Si Li2, Qinchang Chen2, Yiyan Song2, Wulin Tan1#, Zhongxing Wang1# 1Department of Anesthesiology, The First Affiliated Hospital of Sun Yat-sen University, Guangzhou, China; 2Zhongshan School of Medicine, Sun Yat-sen University, Guangzhou, China. *Equal contributors and co-first au- thors. #Equal contributors. Received May 31, 2018; Accepted August 4, 2018; Epub March 15, 2019; Published March 30, 2019 Abstract: Background: Coronary artery disease has been the most common cause of death and the prognosis still needs further improving. Differences in the incidence and prognosis of male and female patients with coronary artery disease have been observed. We constructed this study hoping to understand those differences at the level of gene expression and to help establish gender-specific therapies. Methods: We downloaded the series matrix file of GSE34198 from the Gene Expression Omnibus database and identified differentially expressed genes between male and female patients. Gene ontology, Kyoto Encyclopedia of Genes and Genomes pathway enrichment analy- sis, and GSEA analysis of differentially expressed genes were performed. The protein-protein interaction network was constructed of the differentially expressed genes and the hub genes were identified. Results: A total of 215 up-regulated genes and 353 down-regulated genes were identified. The differentially expressed pathways were mainly related to the function of ribosomes, virus, and related immune response as well as the cell growth and proliferation. -

The DNA Sequence and Comparative Analysis of Human Chromosome 20
articles The DNA sequence and comparative analysis of human chromosome 20 P. Deloukas, L. H. Matthews, J. Ashurst, J. Burton, J. G. R. Gilbert, M. Jones, G. Stavrides, J. P. Almeida, A. K. Babbage, C. L. Bagguley, J. Bailey, K. F. Barlow, K. N. Bates, L. M. Beard, D. M. Beare, O. P. Beasley, C. P. Bird, S. E. Blakey, A. M. Bridgeman, A. J. Brown, D. Buck, W. Burrill, A. P. Butler, C. Carder, N. P. Carter, J. C. Chapman, M. Clamp, G. Clark, L. N. Clark, S. Y. Clark, C. M. Clee, S. Clegg, V. E. Cobley, R. E. Collier, R. Connor, N. R. Corby, A. Coulson, G. J. Coville, R. Deadman, P. Dhami, M. Dunn, A. G. Ellington, J. A. Frankland, A. Fraser, L. French, P. Garner, D. V. Grafham, C. Grif®ths, M. N. D. Grif®ths, R. Gwilliam, R. E. Hall, S. Hammond, J. L. Harley, P. D. Heath, S. Ho, J. L. Holden, P. J. Howden, E. Huckle, A. R. Hunt, S. E. Hunt, K. Jekosch, C. M. Johnson, D. Johnson, M. P. Kay, A. M. Kimberley, A. King, A. Knights, G. K. Laird, S. Lawlor, M. H. Lehvaslaiho, M. Leversha, C. Lloyd, D. M. Lloyd, J. D. Lovell, V. L. Marsh, S. L. Martin, L. J. McConnachie, K. McLay, A. A. McMurray, S. Milne, D. Mistry, M. J. F. Moore, J. C. Mullikin, T. Nickerson, K. Oliver, A. Parker, R. Patel, T. A. V. Pearce, A. I. Peck, B. J. C. T. Phillimore, S. R. Prathalingam, R. W. Plumb, H. Ramsay, C. M. -

Genome-Wide Pooling Approach Identifies SPATA5 As A
European Journal of Human Genetics (2012) 20, 326–332 & 2012 Macmillan Publishers Limited All rights reserved 1018-4813/12 www.nature.com/ejhg ARTICLE Genome-wide pooling approach identifies SPATA5 as a new susceptibility locus for alopecia areata Lina M Forstbauer1,20, Felix F Brockschmidt1,2,20, Valentina Moskvina3, Christine Herold4, Silke Redler1, Alexandra Herzog1, Axel M Hillmer5, Christian Meesters4,6, Stefanie Heilmann1,2, Florian Albert1, Margrieta Alblas1,2, Sandra Hanneken7, Sibylle Eigelshoven7, Kathrin A Giehl8, Dagny Jagielska1,9, Ulrike Blume-Peytavi9, Natalie Garcia Bartels9, Jennifer Kuhn10,11,12, Hans Christian Hennies10,11,12, Matthias Goebeler13, Andreas Jung13, Wiebke K Peitsch14, Anne-Katrin Kortu¨m15, Ingrid Moll15, Roland Kruse16, Gerhard Lutz17, Hans Wolff7, Bettina Blaumeiser18, Markus Bo¨hm19, George Kirov3, Tim Becker4,6, Markus M No¨then1,2 and Regina C Betz*,1 Alopecia areata (AA) is a common hair loss disorder, which is thought to be a tissue-specific autoimmune disease. Previous research has identified a few AA susceptibility genes, most of which are implicated in autoimmunity. To identify new genetic variants and further elucidate the genetic basis of AA, we performed a genome-wide association study using the strategy of pooled DNA genotyping (729 cases, 656 controls). The strongest association was for variants in the HLA region, which confirms the validity of the pooling strategy. The selected top 61 single-nucleotide polymorphisms (SNPs) were analyzed in an independent replication sample (454 cases, 1364 controls). Only one SNP outside of the HLA region (rs304650) showed significant association. This SNP was then analyzed in a second independent replication sample (537 cases, 657 controls). -

Promoter Chip-Chip Analysis in Mouse Testis Reveals Y Chromosome Occupancy by HSF2
Promoter ChIP-chip analysis in mouse testis reveals Y chromosome occupancy by HSF2 Malin Åkerfelt*†‡, Eva Henriksson*†‡, Asta Laiho*, Anniina Vihervaara*†, Karoliina Rautoma*†, Noora Kotaja§ and Lea Sistonen*†¶ *Turku Centre for Biotechnology, University of Turku and Åbo Akademi University, FIN-20520 Turku, Finland; †Department of Biology, Åbo Akademi University, FIN-20520 Turku, Finland; and §Department of Physiology, University of Turku, FIN-20520, Turku, Finland Edited by David C. Page, Massachusetts Institute of Technology, Cambridge, MA, and approved May 21, 2008 (received for review January 21, 2008) The mammalian Y chromosome is essential for spermatogenesis, High-resolution chromatin immunoprecipitation on microarray which is characterized by sperm cell differentiation and chromatin (ChIP-chip) screens have successfully been used for identifying condensation for acquisition of correct shape of the sperm. Dele- direct target genes for many transcription factors (10). For example, tions of the male-specific region of the mouse Y chromosome long Ϸ3% of the genomic loci were found to be targets for HSF in arm (MSYq), harboring multiple copies of a few genes, lead to Saccharomyces cerevisiae and Drosophila exposed to heat stress (11, sperm head defects and impaired fertility. Using chromatin immu- 12). In mammals, however, the existence of three differently noprecipitation on promoter microarray (ChIP-chip) on mouse expressed HSFs (HSF1, HSF2, and HSF4) requires a strategy testis, we found a striking in vivo MSYq occupancy by heat shock to investigate each HSF in a tissue-specific manner. Here, we factor 2 (HSF2), a transcription factor involved in spermatogenesis. chose to dissect the specific role for HSF2 in spermato- HSF2 was also found to regulate the transcription of MSYq resident genesis and to map the in vivo targets for HSF2, by using mouse genes, whose transcriptional regulation has been unknown. -

Tepzz 8Z6z54a T
(19) TZZ ZZ_T (11) EP 2 806 054 A1 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 26.11.2014 Bulletin 2014/48 C40B 40/06 (2006.01) C12Q 1/68 (2006.01) C40B 30/04 (2006.01) C07H 21/00 (2006.01) (21) Application number: 14175049.7 (22) Date of filing: 28.05.2009 (84) Designated Contracting States: (74) Representative: Irvine, Jonquil Claire AT BE BG CH CY CZ DE DK EE ES FI FR GB GR HGF Limited HR HU IE IS IT LI LT LU LV MC MK MT NL NO PL 140 London Wall PT RO SE SI SK TR London EC2Y 5DN (GB) (30) Priority: 28.05.2008 US 56827 P Remarks: •Thecomplete document including Reference Tables (62) Document number(s) of the earlier application(s) in and the Sequence Listing can be downloaded from accordance with Art. 76 EPC: the EPO website 09753364.0 / 2 291 553 •This application was filed on 30-06-2014 as a divisional application to the application mentioned (71) Applicant: Genomedx Biosciences Inc. under INID code 62. Vancouver, British Columbia V6J 1J8 (CA) •Claims filed after the date of filing of the application/ after the date of receipt of the divisional application (72) Inventor: Davicioni, Elai R.68(4) EPC). Vancouver British Columbia V6J 1J8 (CA) (54) Systems and methods for expression- based discrimination of distinct clinical disease states in prostate cancer (57) A system for expression-based discrimination of distinct clinical disease states in prostate cancer is provided that is based on the identification of sets of gene transcripts, which are characterized in that changes in expression of each gene transcript within a set of gene transcripts can be correlated with recurrent or non- recur- rent prostate cancer. -

Evolutionary History of Novel Genes on the Tammar Wallaby Y Chromosome: Implications for Sex Chromosome Evolution
Downloaded from genome.cshlp.org on October 5, 2021 - Published by Cold Spring Harbor Laboratory Press Evolutionary history of novel genes on the tammar wallaby Y chromosome: implications for sex chromosome evolution Veronica J. Murtagh1,2, Denis O’Meally1,9, Natasha Sankovic1,2,3, Margaret L. Delbridge1,2, Yoko Kuroki4, Jeffrey L. Boore5, Atsushi Toyoda6, Kristen S. Jordan1, Andrew J. Pask2,3,7, Marilyn B. Renfree2,3, Asao Fujiyama6,8, Jennifer A. Marshall Graves1,2,3 and Paul D. Waters1,2§ 1 Evolution, Ecology and Genetics, Research School of Biology, The Australian National University, Canberra, ACT 2601, Australia 2 ARC Centre of Excellence for Kangaroo Genomics 3 Department of Zoology, The University of Melbourne, Victoria 3010, Australia 4 RIKEN Research Center for Allergy and Immunology, Immunogenomics, Yokohama, Japan 5 Genome Project Solutions, 1024 Promenade Street, Hercules, California, 94547 USA and DOE Joint Genome Institute, 2800 Mitchell Drive, Walnut Creek, CA 94598 USA 6 National Institute of Genetics, Mishima, Japan 7 Department of Molecular and Cellular Biology, University of Connecticut, Storrs, Connecticut 06260, USA 8 National Institute of Informatics, Tokyo, Japan 9 Institute for Applied Ecology, University of Canberra, ACT 2601, Australia §Corresponding author Evolution, Ecology and Genetics, Research School of Biology, The Australian National University, GPO Box 475, ACT 2601 Canberra, Australia e-mail: [email protected] phone: +61-2-6125 2371 fax: +61-2-6125 8525 - 1 - Downloaded from genome.cshlp.org on October 5, 2021 - Published by Cold Spring Harbor Laboratory Press keywords: Y chromosome, Comparative genomics, Marsupials, X chromosome - 2 - Downloaded from genome.cshlp.org on October 5, 2021 - Published by Cold Spring Harbor Laboratory Press Abstract We report here the isolation and sequencing of ten Y-specific tammar wallaby (Macropus eugenii) BAC clones, revealing five hitherto undescribed tammar wallaby Y genes (in addition to the five genes already described) and several pseudogenes. -

Supplemental Table 3 Two-Class Paired Significance Analysis of Microarrays Comparing Gene Expression Between Paired
Supplemental Table 3 Two‐class paired Significance Analysis of Microarrays comparing gene expression between paired pre‐ and post‐transplant kidneys biopsies (N=8). Entrez Fold q‐value Probe Set ID Gene Symbol Unigene Name Score Gene ID Difference (%) Probe sets higher expressed in post‐transplant biopsies in paired analysis (N=1871) 218870_at 55843 ARHGAP15 Rho GTPase activating protein 15 7,01 3,99 0,00 205304_s_at 3764 KCNJ8 potassium inwardly‐rectifying channel, subfamily J, member 8 6,30 4,50 0,00 1563649_at ‐‐ ‐‐ ‐‐ 6,24 3,51 0,00 1567913_at 541466 CT45‐1 cancer/testis antigen CT45‐1 5,90 4,21 0,00 203932_at 3109 HLA‐DMB major histocompatibility complex, class II, DM beta 5,83 3,20 0,00 204606_at 6366 CCL21 chemokine (C‐C motif) ligand 21 5,82 10,42 0,00 205898_at 1524 CX3CR1 chemokine (C‐X3‐C motif) receptor 1 5,74 8,50 0,00 205303_at 3764 KCNJ8 potassium inwardly‐rectifying channel, subfamily J, member 8 5,68 6,87 0,00 226841_at 219972 MPEG1 macrophage expressed gene 1 5,59 3,76 0,00 203923_s_at 1536 CYBB cytochrome b‐245, beta polypeptide (chronic granulomatous disease) 5,58 4,70 0,00 210135_s_at 6474 SHOX2 short stature homeobox 2 5,53 5,58 0,00 1562642_at ‐‐ ‐‐ ‐‐ 5,42 5,03 0,00 242605_at 1634 DCN decorin 5,23 3,92 0,00 228750_at ‐‐ ‐‐ ‐‐ 5,21 7,22 0,00 collagen, type III, alpha 1 (Ehlers‐Danlos syndrome type IV, autosomal 201852_x_at 1281 COL3A1 dominant) 5,10 8,46 0,00 3493///3 IGHA1///IGHA immunoglobulin heavy constant alpha 1///immunoglobulin heavy 217022_s_at 494 2 constant alpha 2 (A2m marker) 5,07 9,53 0,00 1 202311_s_at