Promoter Chip-Chip Analysis in Mouse Testis Reveals Y Chromosome Occupancy by HSF2
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Small Cell Ovarian Carcinoma: Genomic Stability and Responsiveness to Therapeutics
Gamwell et al. Orphanet Journal of Rare Diseases 2013, 8:33 http://www.ojrd.com/content/8/1/33 RESEARCH Open Access Small cell ovarian carcinoma: genomic stability and responsiveness to therapeutics Lisa F Gamwell1,2, Karen Gambaro3, Maria Merziotis2, Colleen Crane2, Suzanna L Arcand4, Valerie Bourada1,2, Christopher Davis2, Jeremy A Squire6, David G Huntsman7,8, Patricia N Tonin3,4,5 and Barbara C Vanderhyden1,2* Abstract Background: The biology of small cell ovarian carcinoma of the hypercalcemic type (SCCOHT), which is a rare and aggressive form of ovarian cancer, is poorly understood. Tumourigenicity, in vitro growth characteristics, genetic and genomic anomalies, and sensitivity to standard and novel chemotherapeutic treatments were investigated in the unique SCCOHT cell line, BIN-67, to provide further insight in the biology of this rare type of ovarian cancer. Method: The tumourigenic potential of BIN-67 cells was determined and the tumours formed in a xenograft model was compared to human SCCOHT. DNA sequencing, spectral karyotyping and high density SNP array analysis was performed. The sensitivity of the BIN-67 cells to standard chemotherapeutic agents and to vesicular stomatitis virus (VSV) and the JX-594 vaccinia virus was tested. Results: BIN-67 cells were capable of forming spheroids in hanging drop cultures. When xenografted into immunodeficient mice, BIN-67 cells developed into tumours that reflected the hypercalcemia and histology of human SCCOHT, notably intense expression of WT-1 and vimentin, and lack of expression of inhibin. Somatic mutations in TP53 and the most common activating mutations in KRAS and BRAF were not found in BIN-67 cells by DNA sequencing. -
Primepcr™Assay Validation Report
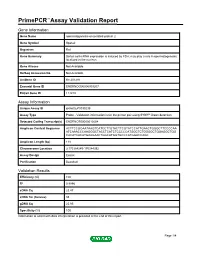
PrimePCR™Assay Validation Report Gene Information Gene Name spermatogenesis-associated protein 2 Gene Symbol Spata2 Organism Rat Gene Summary Sertoli cell mRNA expression is induced by FSH; may play a role in spermatogenesis; localized to the nucleus Gene Aliases Not Available RefSeq Accession No. Not Available UniGene ID Rn.201291 Ensembl Gene ID ENSRNOG00000009207 Entrez Gene ID 114210 Assay Information Unique Assay ID qRnoCEP0030238 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Coding Transcript(s) ENSRNOT00000012604 Amplicon Context Sequence ACTTCCGGAATAAGTCATCCTTGTACTTCGTATCCATTGAACTGGGCTTCCCCAA ATCAAACCCAAGGGCTACCTCATCTCCCCCATGGCTCTGGGGCTGGAGGCTGG CACATCACATGAAGAACTGGCATGGTGCCCATGGACCAGC Amplicon Length (bp) 118 Chromosome Location 3:170384245-170384392 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 100 R2 0.9996 cDNA Cq 23.47 cDNA Tm (Celsius) 85 gDNA Cq 25.95 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report Spata2, Rat Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. Detected Coding Transcript(s) This is a list of the Ensembl transcript ID(s) that this assay will detect. -

PRODUCT SPECIFICATION Prest Antigen ACRV1 Product
PrEST Antigen ACRV1 Product Datasheet PrEST Antigen PRODUCT SPECIFICATION Product Name PrEST Antigen ACRV1 Product Number APrEST80590 Gene Description acrosomal vesicle protein 1 Alternative Gene D11S4365, SP-10, SPACA2 Names Corresponding Anti-ACRV1 (HPA038718) Antibodies Description Recombinant protein fragment of Human ACRV1 Amino Acid Sequence Recombinant Protein Epitope Signature Tag (PrEST) antigen sequence: TSSQPNELSGSIDHQTSVQQLPGEFFSLENPSDAEALYETSSGLNTLSEH GSSEHGSSKHTVAEHTSGEHAE Fusion Tag N-terminal His6ABP (ABP = Albumin Binding Protein derived from Streptococcal Protein G) Expression Host E. coli Purification IMAC purification Predicted MW 25 kDa including tags Usage Suitable as control in WB and preadsorption assays using indicated corresponding antibodies. Purity >80% by SDS-PAGE and Coomassie blue staining Buffer PBS and 1M Urea, pH 7.4. Unit Size 100 µl Concentration Lot dependent Storage Upon delivery store at -20°C. Avoid repeated freeze/thaw cycles. Notes Gently mix before use. Optimal concentrations and conditions for each application should be determined by the user. Product of Sweden. For research use only. Not intended for pharmaceutical development, diagnostic, therapeutic or any in vivo use. No products from Atlas Antibodies may be resold, modified for resale or used to manufacture commercial products without prior written approval from Atlas Antibodies AB. Warranty: The products supplied by Atlas Antibodies are warranted to meet stated product specifications and to conform to label descriptions when used and stored properly. Unless otherwise stated, this warranty is limited to one year from date of sales for products used, handled and stored according to Atlas Antibodies AB's instructions. Atlas Antibodies AB's sole liability is limited to replacement of the product or refund of the purchase price. -

TCTE1 Is a Conserved Component of the Dynein Regulatory Complex and Is Required for Motility and Metabolism in Mouse Spermatozoa
TCTE1 is a conserved component of the dynein regulatory complex and is required for motility and metabolism in mouse spermatozoa Julio M. Castanedaa,b,1, Rong Huac,d,1, Haruhiko Miyatab, Asami Ojib,e, Yueshuai Guoc,d, Yiwei Chengc,d, Tao Zhouc,d, Xuejiang Guoc,d, Yiqiang Cuic,d, Bin Shenc, Zibin Wangc, Zhibin Huc,f, Zuomin Zhouc,d, Jiahao Shac,d, Renata Prunskaite-Hyyrylainena,g,h, Zhifeng Yua,i, Ramiro Ramirez-Solisj, Masahito Ikawab,e,k,2, Martin M. Matzuka,g,i,l,m,n,2, and Mingxi Liuc,d,2 aDepartment of Pathology and Immunology, Baylor College of Medicine, Houston, TX 77030; bResearch Institute for Microbial Diseases, Osaka University, Suita, Osaka 5650871, Japan; cState Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing 210029, People’s Republic of China; dDepartment of Histology and Embryology, Nanjing Medical University, Nanjing 210029, People’s Republic of China; eGraduate School of Pharmaceutical Sciences, Osaka University, Suita, Osaka 5650871, Japan; fAnimal Core Facility of Nanjing Medical University, Nanjing 210029, People’s Republic of China; gCenter for Reproductive Medicine, Baylor College of Medicine, Houston, TX 77030; hFaculty of Biochemistry and Molecular Medicine, University of Oulu, Oulu FI-90014, Finland; iCenter for Drug Discovery, Baylor College of Medicine, Houston, TX 77030; jWellcome Trust Sanger Institute, Hinxton CB10 1SA, United Kingdom; kThe Institute of Medical Science, The University of Tokyo, Minato-ku, Tokyo 1088639, Japan; lDepartment of Molecular and Cellular Biology, Baylor College of Medicine, Houston, TX 77030; mDepartment of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030; and nDepartment of Pharmacology, Baylor College of Medicine, Houston, TX 77030 Contributed by Martin M. -

ACRV1 (NM 020069) Human Tagged ORF Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC214263 ACRV1 (NM_020069) Human Tagged ORF Clone Product data: Product Type: Expression Plasmids Product Name: ACRV1 (NM_020069) Human Tagged ORF Clone Tag: Myc-DDK Symbol: ACRV1 Synonyms: D11S4365; SP-10; SPACA2 Vector: pCMV6-Entry (PS100001) E. coli Selection: Kanamycin (25 ug/mL) Cell Selection: Neomycin ORF Nucleotide >RC214263 representing NM_020069 Sequence: Red=Cloning site Blue=ORF Green=Tags(s) TTTTGTAATACGACTCACTATAGGGCGGCCGGGAATTCGTCGACTGGATCCGGTACCGAGGAGATCTGCC GCCGCGATCGCC ATGAACAGGTTTCTCTTGCTAATGAGTCTTTATCTGCTTGGATCTGCCAGAGGAACATCAAGTCAGCCTA ATGAGCTTTCTGGCTCCATAGATCATCAAACTTCAGTTCAGCAACTTCCAGGTGAGTTCTTTTCACTTGA AAACCCTTCTGATGCTGAGGCTTTATATGAGACTTCTTCAGGCCTGAACACTTTAAGTGAGCATGGTTCC AGTGAGCATGGTTCAAGCAAGCACACTGTGGCCGAGCACACTTCTGGAGAACATGCTGAGAGTGAGCATG CTTCAGGTGAGCCCGCTGCGACTGAACATGCTGAAGGTGAGCATACTGTAGGTGAGCAGCCTTCAGGAGA ACAGCCTTCAGGTGAACACCTCTCCGGAGAACAGCCTTTGAGTGAGCTTGAGTCAGGTGAACAGCCTTCA GATGAACAGCCTTCAGGTGAACATGGCTCCGGTGAACAGCCTTCTGGTGAGCAGGCCTCGGGTGAACAGC CTTCAGGCACAATATTAAATTGCTACACATGTGCTTATATGAATGATCAAGGAAAATGTCTTCGTGGAGA GGGAACCTGCATCACTCAGAATTCCCAGCAGTGCATGTTAAAGAAGATCTTTGAAGGTGGAAAACTCCAA TTCATGGTTCAAGGGTGTGAGAACATGTGCCCATCTATGAACCTCTTCTCCCATGGAACGAGGATGCAAA TTATATGCTGTCGAAATCAATCTTTCTGCAATAAGATC ACGCGTACGCGGCCGCTCGAGCAGAAACTCATCTCAGAAGAGGATCTGGCAGCAAATGATATCCTGGATT ACAAGGATGACGACGATAAGGTTTAA This product is -

1 Retrotransposons and Pseudogenes Regulate Mrnas and Lncrnas Via the Pirna Pathway 1 in the Germline 2 3 Toshiaki Watanabe*, E
Downloaded from genome.cshlp.org on October 6, 2021 - Published by Cold Spring Harbor Laboratory Press 1 Retrotransposons and pseudogenes regulate mRNAs and lncRNAs via the piRNA pathway 2 in the germline 3 4 Toshiaki Watanabe*, Ee-chun Cheng, Mei Zhong, and Haifan Lin* 5 Yale Stem Cell Center and Department of Cell Biology, Yale University School of Medicine, New Haven, 6 Connecticut 06519, USA 7 8 Running Title: Pachytene piRNAs regulate mRNAs and lncRNAs 9 10 Key Words: retrotransposon, pseudogene, lncRNA, piRNA, Piwi, spermatogenesis 11 12 *Correspondence: [email protected]; [email protected] 13 1 Downloaded from genome.cshlp.org on October 6, 2021 - Published by Cold Spring Harbor Laboratory Press 14 ABSTRACT 15 The eukaryotic genome has vast intergenic regions containing transposons, pseudogenes, and other 16 repetitive sequences. They produce numerous long non-coding RNAs (lncRNAs) and PIWI-interacting 17 RNAs (piRNAs), yet the functions of the vast intergenic regions remain largely unknown. Mammalian 18 piRNAs are abundantly expressed in late spermatocytes and round spermatids, coinciding with the 19 widespread expression of lncRNAs in these cells. Here, we show that piRNAs derived from transposons 20 and pseudogenes mediate the degradation of a large number of mRNAs and lncRNAs in mouse late 21 spermatocytes. In particular, they have a large impact on the lncRNA transcriptome, as a quarter of 22 lncRNAs expressed in late spermatocytes are up-regulated in mice deficient in the piRNA pathway. 23 Furthermore, our genomic and in vivo functional analyses reveal that retrotransposon sequences in the 24 3´UTR of mRNAs are targeted by piRNAs for degradation. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Genetics of Amyotrophic Lateral Sclerosis in the Han Chinese
Genetics of amyotrophic lateral sclerosis in the Han Chinese Ji He A thesis submitted for the degree of Master of Philosophy at The University of Queensland in 2015 The University of Queensland Diamantina Institute 1 Abstract Amyotrophic lateral sclerosis is the most frequently occurring neuromuscular degenerative disorders, and has an obscure aetiology. Whilst major progress has been made, the majority of the genetic variation involved in ALS is, as yet, undefined. In this thesis, multiple genetic studies have been conducted to advance our understanding of the genetic architecture of the disease. In the light of the paucity of comprehensive genetic studies performed in Chinese, the presented study focused on advancing our current understanding in genetics of ALS in the Han Chinese population. To identify genetic variants altering risk of ALS, a genome-wide association study (GWAS) was performed. The study included 1,324 Chinese ALS cases and 3,115 controls. After quality control, a number of analyses were performed in a cleaned dataset of 1,243 cases and 2,854 controls that included: a genome-wide association analysis to identify SNPs associated with ALS; a genomic restricted maximum likelihood (GREML) analysis to estimate the proportion of the phenotypic variance in ALS liability due to common SNPs; and a gene- based analysis to identify genes associated with ALS. There were no genome-wide significant SNPs or genes associated with ALS. However, it was estimated that 17% (SE: 0.05; P=6×10-5) of the phenotypic variance in ALS liability was due to common SNPs. The top associated SNP was within GNAS (rs4812037; p =7×10-7). -

Molecular Classification of Patients with Unexplained Hamartomatous and Hyperplastic Polyposis
ORIGINAL CONTRIBUTION Molecular Classification of Patients With Unexplained Hamartomatous and Hyperplastic Polyposis Kevin Sweet, MS, CGC Context Significant proportions of patients with hamartomatous polyposis or with Joseph Willis, MD hyperplastic/mixed polyposis remain without specific clinical and molecular diagnosis Xiao-Ping Zhou, MD, PhD or present atypically. Assigning a syndromic diagnosis is important because it guides management, especially surveillance and prophylactic surgery. Carol Gallione, PhD Objective To systematically classify patients with unexplained hamartomatous or hy- Takeshi Sawada, MD, PhD perplastic/mixed polyposis by extensive molecular analysis in the context of central Pia Alhopuro, MD rereview of histopathology results. Sok Kean Khoo, PhD Design, Setting, and Patients Prospective, referral-based study of 49 unrelated patients from outside institutions (n=28) and at a comprehensive cancer center (n=21), Attila Patocs, MD, PhD conducted from May 2, 2002, until December 15, 2004. Germline analysis of PTEN, Cossette Martin, PhD BMPR1A, STK11 (sequence, deletion), SMAD4, and ENG (sequence), specific exon screen- Scott Bridgeman, BSc ing of BRAF, MYH, and BHD, and rereview of polyp histology results were performed. John Heinz, PhD Main Outcome Measures Molecular, clinical, and histopathological findings in pa- tients with unexplained polyposis. Robert Pilarski, MS, CGC Results Of the 49 patients, 11 (22%) had germline mutations. Of 14 patients with Rainer Lehtonen, BSc juvenile polyposis, 2 with early-onset disease had mutations in ENG, encoding endo- Thomas W. Prior, PhD glin, previously only associated with hereditary hemorrhagic telangiectasia; 1 had hemi- zygous deletion encompassing PTEN and BMPR1A; and 1 had an SMAD4 mutation. Thierry Frebourg, MD, PhD One individual previously classified with Peutz-Jeghers syndrome had a PTEN dele- Bin Tean Teh, MD, PhD tion. -

Mouse Spata2 Conditional Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Spata2 Conditional Knockout Project (CRISPR/Cas9) Objective: To create a Spata2 conditional knockout Mouse model (C57BL/6J) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Spata2 gene (NCBI Reference Sequence: NM_170756 ; Ensembl: ENSMUSG00000047030 ) is located on Mouse chromosome 2. 3 exons are identified, with the ATG start codon in exon 2 and the TAG stop codon in exon 3 (Transcript: ENSMUST00000057627). Exon 2~3 will be selected as conditional knockout region (cKO region). Deletion of this region should result in the loss of function of the Mouse Spata2 gene. To engineer the targeting vector, homologous arms and cKO region will be generated by PCR using BAC clone RP24-144E24 as template. Cas9, gRNA and targeting vector will be co-injected into fertilized eggs for cKO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Homozygous knockout leads to small testes, oligospermia, asthenozoospermia, reduced male fertility and decreased male germ cell numbers. It also affects necroptosis and increases inflammatory responses. Exon 2~3 covers 100.0% of the coding region. Start codon is in exon 2, and stop codon is in exon 3. The size of intron 1 for 5'-loxP site insertion: 7165 bp. The size of effective cKO region: ~2396 bp. The cKO region does not have any other known gene. Page 1 of 8 https://www.alphaknockout.com Overview of the Targeting Strategy gRNA region Wildtype allele T A 5' gRNA region G 3' 1 2 3 Targeting vector T A G Targeted allele T A G Constitutive KO allele (After Cre recombination) Legends Exon of mouse Spata2 Homology arm cKO region loxP site Page 2 of 8 https://www.alphaknockout.com Overview of the Dot Plot Window size: 10 bp Forward Reverse Complement Sequence 12 Note: The sequence of homologous arms and cKO region is aligned with itself to determine if there are tandem repeats. -

Mouse Acrv1 Conditional Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Acrv1 Conditional Knockout Project (CRISPR/Cas9) Objective: To create a Acrv1 conditional knockout Mouse model (C57BL/6J) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Acrv1 gene (NCBI Reference Sequence: NM_007391 ; Ensembl: ENSMUSG00000032110 ) is located on Mouse chromosome 9. 4 exons are identified, with the ATG start codon in exon 1 and the TAG stop codon in exon 4 (Transcript: ENSMUST00000034620). Exon 2~3 will be selected as conditional knockout region (cKO region). Deletion of this region should result in the loss of function of the Mouse Acrv1 gene. To engineer the targeting vector, homologous arms and cKO region will be generated by PCR using BAC clone RP23-339F21 as template. Cas9, gRNA and targeting vector will be co-injected into fertilized eggs for cKO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Exon 2~3 is not frameshift exon, and covers 78.16% of the coding region. The size of intron 1 for 5'-loxP site insertion: 847 bp, and the size of intron 3 for 3'-loxP site insertion: 1789 bp. The size of effective cKO region: ~2999 bp. The cKO region does not have any other known gene. Page 1 of 7 https://www.alphaknockout.com Overview of the Targeting Strategy Wildtype allele 5' gRNA region gRNA region 3' 1 2 3 4 Targeting vector Targeted allele Constitutive KO allele (After Cre recombination) Legends Homology arm Exon of mouse Acrv1 cKO region loxP site Page 2 of 7 https://www.alphaknockout.com Overview of the Dot Plot Window size: 10 bp Forward Reverse Complement Sequence 12 Note: The sequence of homologous arms and cKO region is aligned with itself to determine if there are tandem repeats. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase