Association of Rs610604 in TNFAIP3 and Rs17728338 in TNIP1 Gene
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Acute Lymphoblastic Leukemia Patient with Variant ATF7IP
ISSN 1941-5923 © Am J Case Rep, 2017; 18: 1204-1208 DOI: 10.12659/AJCR.906300 Received: 2017.07.20 Accepted: 2017.08.07 Acute Lymphoblastic Leukemia Patient with Published: 2017.11.14 Variant ATF7IP/PDGFRB Fusion and Favorable Response to Tyrosine Kinase Inhibitor Treatment: A Case Report Authors’ Contribution: BCDF 1,2 Ge Zhang* 1 Department of Pediatrics, Key Laboratory of Birth Defects and Related Diseases Study Design A BCD 1 Yanle Zhang* of Women and Children, Ministry of Health, West China Second University Data Collection B Hospital, Sichuan University, Chengdu, Sichuan, P.R. China Statistical Analysis C BD 1 Jianrong Wu 2 Department of Laboratory Medicine, West China Second University Hospital, Data Interpretation D AE 3 Yan Chen Sichuan University, Chengdu, Sichuan, P.R. China Manuscript Preparation E AE 1 Zhigui Ma 3 Department of Pediatrics, Affiliated Hospital of Zunyi Medical College, Zunyi, Literature Search F Guizhou, P.R. China Funds Collection G * Ge Zhang and Yanle Zhang contributed equally to this work Corresponding Authors: Zhigui Ma, e-mail: [email protected]; Yan Chen, e-mail: [email protected] Conflict of interest: None declared Patient: Female, 14-month-old Final Diagnosis: Acute lymphoblastic leukemia Symptoms: Fever Medication: — Clinical Procedure: — Specialty: Hematology Objective: Rare disease Background: Chromosomal translocations involving the PDGFRB gene have been reported in a broad spectrum of hemato- logical malignancies. An ATF7IP/PDGFRB fusion was recently identified in a Philadelphia chromosome-like (Ph- like) B-progenitor acute lymphoblastic leukemia (B-ALL) patient. Here we report on a special case of a Ph-like ALL patient who had a variant ATF7IP/PDGFRB fusion. -

Small Cell Ovarian Carcinoma: Genomic Stability and Responsiveness to Therapeutics
Gamwell et al. Orphanet Journal of Rare Diseases 2013, 8:33 http://www.ojrd.com/content/8/1/33 RESEARCH Open Access Small cell ovarian carcinoma: genomic stability and responsiveness to therapeutics Lisa F Gamwell1,2, Karen Gambaro3, Maria Merziotis2, Colleen Crane2, Suzanna L Arcand4, Valerie Bourada1,2, Christopher Davis2, Jeremy A Squire6, David G Huntsman7,8, Patricia N Tonin3,4,5 and Barbara C Vanderhyden1,2* Abstract Background: The biology of small cell ovarian carcinoma of the hypercalcemic type (SCCOHT), which is a rare and aggressive form of ovarian cancer, is poorly understood. Tumourigenicity, in vitro growth characteristics, genetic and genomic anomalies, and sensitivity to standard and novel chemotherapeutic treatments were investigated in the unique SCCOHT cell line, BIN-67, to provide further insight in the biology of this rare type of ovarian cancer. Method: The tumourigenic potential of BIN-67 cells was determined and the tumours formed in a xenograft model was compared to human SCCOHT. DNA sequencing, spectral karyotyping and high density SNP array analysis was performed. The sensitivity of the BIN-67 cells to standard chemotherapeutic agents and to vesicular stomatitis virus (VSV) and the JX-594 vaccinia virus was tested. Results: BIN-67 cells were capable of forming spheroids in hanging drop cultures. When xenografted into immunodeficient mice, BIN-67 cells developed into tumours that reflected the hypercalcemia and histology of human SCCOHT, notably intense expression of WT-1 and vimentin, and lack of expression of inhibin. Somatic mutations in TP53 and the most common activating mutations in KRAS and BRAF were not found in BIN-67 cells by DNA sequencing. -
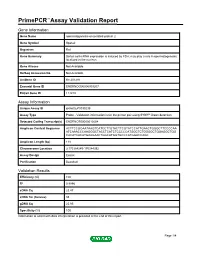
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name spermatogenesis-associated protein 2 Gene Symbol Spata2 Organism Rat Gene Summary Sertoli cell mRNA expression is induced by FSH; may play a role in spermatogenesis; localized to the nucleus Gene Aliases Not Available RefSeq Accession No. Not Available UniGene ID Rn.201291 Ensembl Gene ID ENSRNOG00000009207 Entrez Gene ID 114210 Assay Information Unique Assay ID qRnoCEP0030238 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Coding Transcript(s) ENSRNOT00000012604 Amplicon Context Sequence ACTTCCGGAATAAGTCATCCTTGTACTTCGTATCCATTGAACTGGGCTTCCCCAA ATCAAACCCAAGGGCTACCTCATCTCCCCCATGGCTCTGGGGCTGGAGGCTGG CACATCACATGAAGAACTGGCATGGTGCCCATGGACCAGC Amplicon Length (bp) 118 Chromosome Location 3:170384245-170384392 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 100 R2 0.9996 cDNA Cq 23.47 cDNA Tm (Celsius) 85 gDNA Cq 25.95 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report Spata2, Rat Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. Detected Coding Transcript(s) This is a list of the Ensembl transcript ID(s) that this assay will detect. -

Laboratory Mouse Models for the Human Genome-Wide Associations
Laboratory Mouse Models for the Human Genome-Wide Associations The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Kitsios, Georgios D., Navdeep Tangri, Peter J. Castaldi, and John P. A. Ioannidis. 2010. Laboratory mouse models for the human genome-wide associations. PLoS ONE 5(11): e13782. Published Version doi:10.1371/journal.pone.0013782 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:8592157 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA Laboratory Mouse Models for the Human Genome-Wide Associations Georgios D. Kitsios1,4, Navdeep Tangri1,6, Peter J. Castaldi1,2,4,5, John P. A. Ioannidis1,2,3,4,5,7,8* 1 Institute for Clinical Research and Health Policy Studies, Tufts Medical Center, Boston, Massachusetts, United States of America, 2 Tufts University School of Medicine, Boston, Massachusetts, United States of America, 3 Department of Hygiene and Epidemiology, University of Ioannina School of Medicine and Biomedical Research Institute, Foundation for Research and Technology-Hellas, Ioannina, Greece, 4 Tufts Clinical and Translational Science Institute, Tufts Medical Center, Boston, Massachusetts, United States of America, 5 Department of Medicine, Center for Genetic Epidemiology and Modeling, Tufts Medical Center, Tufts University -

Genetics of Amyotrophic Lateral Sclerosis in the Han Chinese
Genetics of amyotrophic lateral sclerosis in the Han Chinese Ji He A thesis submitted for the degree of Master of Philosophy at The University of Queensland in 2015 The University of Queensland Diamantina Institute 1 Abstract Amyotrophic lateral sclerosis is the most frequently occurring neuromuscular degenerative disorders, and has an obscure aetiology. Whilst major progress has been made, the majority of the genetic variation involved in ALS is, as yet, undefined. In this thesis, multiple genetic studies have been conducted to advance our understanding of the genetic architecture of the disease. In the light of the paucity of comprehensive genetic studies performed in Chinese, the presented study focused on advancing our current understanding in genetics of ALS in the Han Chinese population. To identify genetic variants altering risk of ALS, a genome-wide association study (GWAS) was performed. The study included 1,324 Chinese ALS cases and 3,115 controls. After quality control, a number of analyses were performed in a cleaned dataset of 1,243 cases and 2,854 controls that included: a genome-wide association analysis to identify SNPs associated with ALS; a genomic restricted maximum likelihood (GREML) analysis to estimate the proportion of the phenotypic variance in ALS liability due to common SNPs; and a gene- based analysis to identify genes associated with ALS. There were no genome-wide significant SNPs or genes associated with ALS. However, it was estimated that 17% (SE: 0.05; P=6×10-5) of the phenotypic variance in ALS liability was due to common SNPs. The top associated SNP was within GNAS (rs4812037; p =7×10-7). -

Modulation of NF-Κb Signalling by Microbial Pathogens
REVIEWS Modulation of NF‑κB signalling by microbial pathogens Masmudur M. Rahman and Grant McFadden Abstract | The nuclear factor-κB (NF‑κB) family of transcription factors plays a central part in the host response to infection by microbial pathogens, by orchestrating the innate and acquired host immune responses. The NF‑κB proteins are activated by diverse signalling pathways that originate from many different cellular receptors and sensors. Many successful pathogens have acquired sophisticated mechanisms to regulate the NF‑κB signalling pathways by deploying subversive proteins or hijacking the host signalling molecules. Here, we describe the mechanisms by which viruses and bacteria micromanage the host NF‑κB signalling circuitry to favour the continued survival of the pathogen. The nuclear factor-κB (NF-κB) family of transcription Signalling targets upstream of NF‑κB factors regulates the expression of hundreds of genes that NF-κB proteins are tightly regulated in both the cyto- are associated with diverse cellular processes, such as pro- plasm and the nucleus6. Under normal physiological liferation, differentiation and death, as well as innate and conditions, NF‑κB complexes remain inactive in the adaptive immune responses. The mammalian NF‑κB cytoplasm through a direct interaction with proteins proteins are members of the Rel domain-containing pro- of the inhibitor of NF-κB (IκB) family, including IκBα, tein family: RELA (also known as p65), RELB, c‑REL, IκBβ and IκBε (also known as NF-κBIα, NF-κBIβ and the NF-κB p105 subunit (also known as NF‑κB1; which NF-κBIε, respectively); IκB proteins mask the nuclear is cleaved into the p50 subunit) and the NF-κB p100 localization domains in the NF‑κB complex, thus subunit (also known as NF‑κB2; which is cleaved into retaining the transcription complex in the cytoplasm. -

Biological and Prognostic Significance of Chromosome 5Q Deletions in Myeloid Malignancies Aristoteles A.N
Review Biological and Prognostic Significance of Chromosome 5q Deletions in Myeloid Malignancies Aristoteles A.N. Giagounidis,1Ulrich Germing,2 and Carlo Aul1 Abstract The presence of del(5q), either as the sole karyotypic abnormality or as part of a more complex karyotype, has distinct clinical implications for myelodysplastic syndromes (MDS) and acute myeloid leukemia. The 5qÀ syndrome, a subtype of low-riskMDS, is characterized by an isolated 5q deletion and <5% blasts in the bone marrow and can serve as a useful model for studying the role of 5q deletions in the pathogenesis and prognosis of myeloid malignancies. Recent clinical results with lenalidomide, an oral immunomodulatory drug, have shown durable erythroid responses, including transfusion independence and complete cytogenetic remissions in patients with del(5q) MDS with or without additional chromosomal abnormalities. These results indicate that lenalidomide can overcome the pathogenic effect of 5q deletion in MDS and restore bone marrow balance. The data provide important new insights into the pathobiology of 5q chromo- somal deletions in myeloid malignancies. Cytogenetic abnormalities are detected in the bone marrow of preponderance, refractory macrocytic anemia, normal or high over 50% of patients diagnosed with primary myelodysplastic platelet counts, hypolobulated megakaryocytes, and modest syndromes (MDS) or myeloid leukemias, and up to 80% of leukopenia (11, 14, 17). The prognosis is favorable in 5qÀ patients with secondary or therapy-related MDS (1, 2). These syndrome with relatively low risk of transformation to AML abnormalities can be characterized as being balanced or (11, 18). Although the limits of 5q deletions vary among unbalanced (3, 4). Balanced cytogenetic abnormalities include patients with 5qÀ syndrome, the most frequent deletion is reciprocal translocations, inversions, and insertions (3, 5, 6). -

Mouse Spata2 Conditional Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Spata2 Conditional Knockout Project (CRISPR/Cas9) Objective: To create a Spata2 conditional knockout Mouse model (C57BL/6J) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Spata2 gene (NCBI Reference Sequence: NM_170756 ; Ensembl: ENSMUSG00000047030 ) is located on Mouse chromosome 2. 3 exons are identified, with the ATG start codon in exon 2 and the TAG stop codon in exon 3 (Transcript: ENSMUST00000057627). Exon 2~3 will be selected as conditional knockout region (cKO region). Deletion of this region should result in the loss of function of the Mouse Spata2 gene. To engineer the targeting vector, homologous arms and cKO region will be generated by PCR using BAC clone RP24-144E24 as template. Cas9, gRNA and targeting vector will be co-injected into fertilized eggs for cKO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Homozygous knockout leads to small testes, oligospermia, asthenozoospermia, reduced male fertility and decreased male germ cell numbers. It also affects necroptosis and increases inflammatory responses. Exon 2~3 covers 100.0% of the coding region. Start codon is in exon 2, and stop codon is in exon 3. The size of intron 1 for 5'-loxP site insertion: 7165 bp. The size of effective cKO region: ~2396 bp. The cKO region does not have any other known gene. Page 1 of 8 https://www.alphaknockout.com Overview of the Targeting Strategy gRNA region Wildtype allele T A 5' gRNA region G 3' 1 2 3 Targeting vector T A G Targeted allele T A G Constitutive KO allele (After Cre recombination) Legends Exon of mouse Spata2 Homology arm cKO region loxP site Page 2 of 8 https://www.alphaknockout.com Overview of the Dot Plot Window size: 10 bp Forward Reverse Complement Sequence 12 Note: The sequence of homologous arms and cKO region is aligned with itself to determine if there are tandem repeats. -

The UBE2L3 Ubiquitin Conjugating Enzyme: Interplay with Inflammasome Signalling and Bacterial Ubiquitin Ligases
The UBE2L3 ubiquitin conjugating enzyme: interplay with inflammasome signalling and bacterial ubiquitin ligases Matthew James George Eldridge 2018 Imperial College London Department of Medicine Submitted to Imperial College London for the degree of Doctor of Philosophy 1 Abstract Inflammasome-controlled immune responses such as IL-1β release and pyroptosis play key roles in antimicrobial immunity and are heavily implicated in multiple hereditary autoimmune diseases. Despite extensive knowledge of the mechanisms regulating inflammasome activation, many downstream responses remain poorly understood or uncharacterised. The cysteine protease caspase-1 is the executor of inflammasome responses, therefore identifying and characterising its substrates is vital for better understanding of inflammasome-mediated effector mechanisms. Using unbiased proteomics, the Shenoy grouped identified the ubiquitin conjugating enzyme UBE2L3 as a target of caspase-1. In this work, I have confirmed UBE2L3 as an indirect target of caspase-1 and characterised its role in inflammasomes-mediated immune responses. I show that UBE2L3 functions in the negative regulation of cellular pro-IL-1 via the ubiquitin- proteasome system. Following inflammatory stimuli, UBE2L3 assists in the ubiquitylation and degradation of newly produced pro-IL-1. However, in response to caspase-1 activation, UBE2L3 is itself targeted for degradation by the proteasome in a caspase-1-dependent manner, thereby liberating an additional pool of IL-1 which may be processed and released. UBE2L3 therefore acts a molecular rheostat, conferring caspase-1 an additional level of control over this potent cytokine, ensuring that it is efficiently secreted only in appropriate circumstances. These findings on UBE2L3 have implications for IL-1- driven pathology in hereditary fever syndromes, and autoinflammatory conditions associated with UBE2L3 polymorphisms. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Efficient Analysis of Mouse Genome Sequences Reveal Many Nonsense Variants
Efficient analysis of mouse genome sequences reveal many nonsense variants Sophie Steelanda,b,1, Steven Timmermansa,b,1, Sara Van Ryckeghema,b, Paco Hulpiaua,b, Yvan Saeysa,c, Marc Van Montagud,e,f,2, Roosmarijn E. Vandenbrouckea,b,3, and Claude Liberta,b,2,3 aInflammation Research Center, Flanders Institute for Biotechnology (VIB), 9052 Ghent, Belgium; bDepartment of Biomedical Molecular Biology, Ghent University, 9052 Ghent, Belgium; cDepartment of Internal Medicine, Ghent University, 9052 Ghent, Belgium; dDepartment of Plant Systems Biology, VIB, 9052 Ghent, Belgium; eDepartment of Plant Biotechnology and Bioinformatics, Ghent University, 9052 Ghent, Belgium; and fInternational Plant Biotechnology Outreach, VIB, Ghent, Belgium Contributed by Marc Van Montagu, March 30, 2016 (sent for review December 31, 2015; reviewed by Bruce Beutler, Stefano Bruscoli, Stefan Rose-John, and Klaus Schulze-Osthoff) Genetic polymorphisms in coding genes play an important role alive, archiving them, and distributing mutant strains to in- when using mouse inbred strains as research models. They have terested users (4). been shown to influence research results, explain phenotypical Since Clarence Little showed in the early 20th century that the differences between inbred strains, and increase the amount of principle of inbreeding also applies to mice, several hundred interesting gene variants present in the many available inbred inbred mouse strains have been generated (5). Some of these lines. SPRET/Ei is an inbred strain derived from Mus spretus that strains display specific phenotypes that are the result of a mutant has ∼1% sequence difference with the C57BL/6J reference ge- gene, and in several cases have formed the basis for identifying nome. -

Genome-Wide Pooling Approach Identifies SPATA5 As a New Susceptibility Locus for Alopecia Areata Regina C
Genome-wide pooling approach identifies SPATA5 as a new susceptibility locus for alopecia areata Regina C. Betz, Lina M. Forstbauer, Felix F. Brockschmidt, Valentina Moskvina, Christine Herold, Silke Redler, Alexandra Herzog, Axel M. Hillmer, Christian Meesters, Stefanie Heilmann, et al. To cite this version: Regina C. Betz, Lina M. Forstbauer, Felix F. Brockschmidt, Valentina Moskvina, Christine Herold, et al.. Genome-wide pooling approach identifies SPATA5 as a new susceptibility locus for alopecia areata. European Journal of Human Genetics, Nature Publishing Group, 2011, 10.1038/ejhg.2011.185. hal- 00691329 HAL Id: hal-00691329 https://hal.archives-ouvertes.fr/hal-00691329 Submitted on 26 Apr 2012 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. 1 Genome-wide pooling approach identifies SPATA5 as a new susceptibility locus for 2 alopecia areata 3 4 Lina M. Forstbauer1*, Felix F. Brockschmidt1,2*, Valentina Moskvina3, Christine 5 Herold4, Silke Redler1, Alexandra Herzog1, Axel M. Hillmer5, Christian Meesters4,6, 6 Stefanie Heilmann1,2, Florian Albert1, Margrieta Alblas1,2, Sandra Hanneken7, Sibylle 7 Eigelshoven7, Kathrin A. Giehl8, Dagny Jagielska1,9, Ulrike Blume-Peytavi9, Natalie 8 Garcia Bartels9, Jennifer Kuhn10,11,12, Hans Christian Hennies10,11,12, Matthias 9 Goebeler13, Andreas Jung13, Wiebke K.